Scientific brief
Conditional Deletion of Large DNA Fragment in iPSCs by ssODN-Mediated Insertion of LoxP Sites without Antibiotic Selection
Abstract
Cre-lox is an essential tool for creating conditional gene knockouts in human induced pluripotent stem cells (hiPSCs) for disease modeling and drug screening. Traditionally, both loxP sites are inserted into specific regions of the genome through homology-directed repair (HDR) prompted by delivery of CRISPR-Cas9 and a single donor template. Here, loxP sites were sequentially inserted to flank exons 45-55 in the dystrophin (DMD) gene. Deletion of this mutational hotspot could treat approximately 47% of all Duchenne muscular dystrophy patients.1 The efficiency of loxP insertion and HDR is dependent on the size of the donor template, and larger cassettes are inherently more challenging to insert. Donor sequences for the Cre-lox system generally include both loxP sites, an antibiotic selection cassette, and the DNA sequence to be deleted, resulting in a large cassette. Hotta et al., used the MaxCyte® ExPERTTM platform to deliver two small single-stranded oligodeoxynucleotides (ssODNs) encoding 34 bp loxP sites into hiPSCs.2 MaxCyte electroporation of ssODNs showed high knockin efficiencies without antibiotic selection which led to the precise deletion of a large 342kb DNA genomic region.
Figure 1. Sequential Electroporation of loxP Sites in hiPSCs.
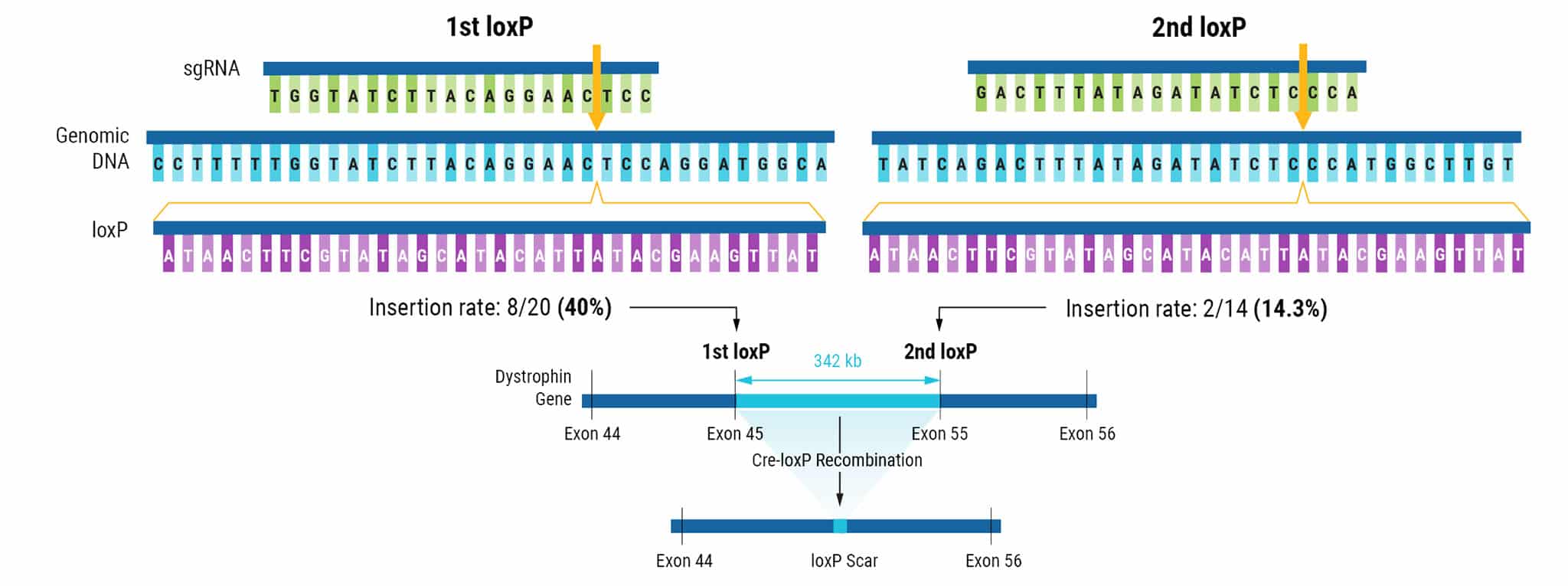
CRISPR-Cas9 RNPs and a 134 bp ssODN donor template were delivered to hiPSCs via electroporation (EP). LoxP sites targeted to the DMD gene flank exons 45-55. The insertion rate for the first loxP site was 40%, with 8 of 20 screened hiPSC clones positive for loxP. After the second electroporation, 2 of 14 clones screened contained the additional loxP site at a frequency of 14.3%. The introduction of Cre recombinase to engineered cells with functional loxP sites resulted in the removal of exons 45-55.
Figure 2. Cre-loxP Recombination Results in the Deletion of 342 kb Genomic DNA.
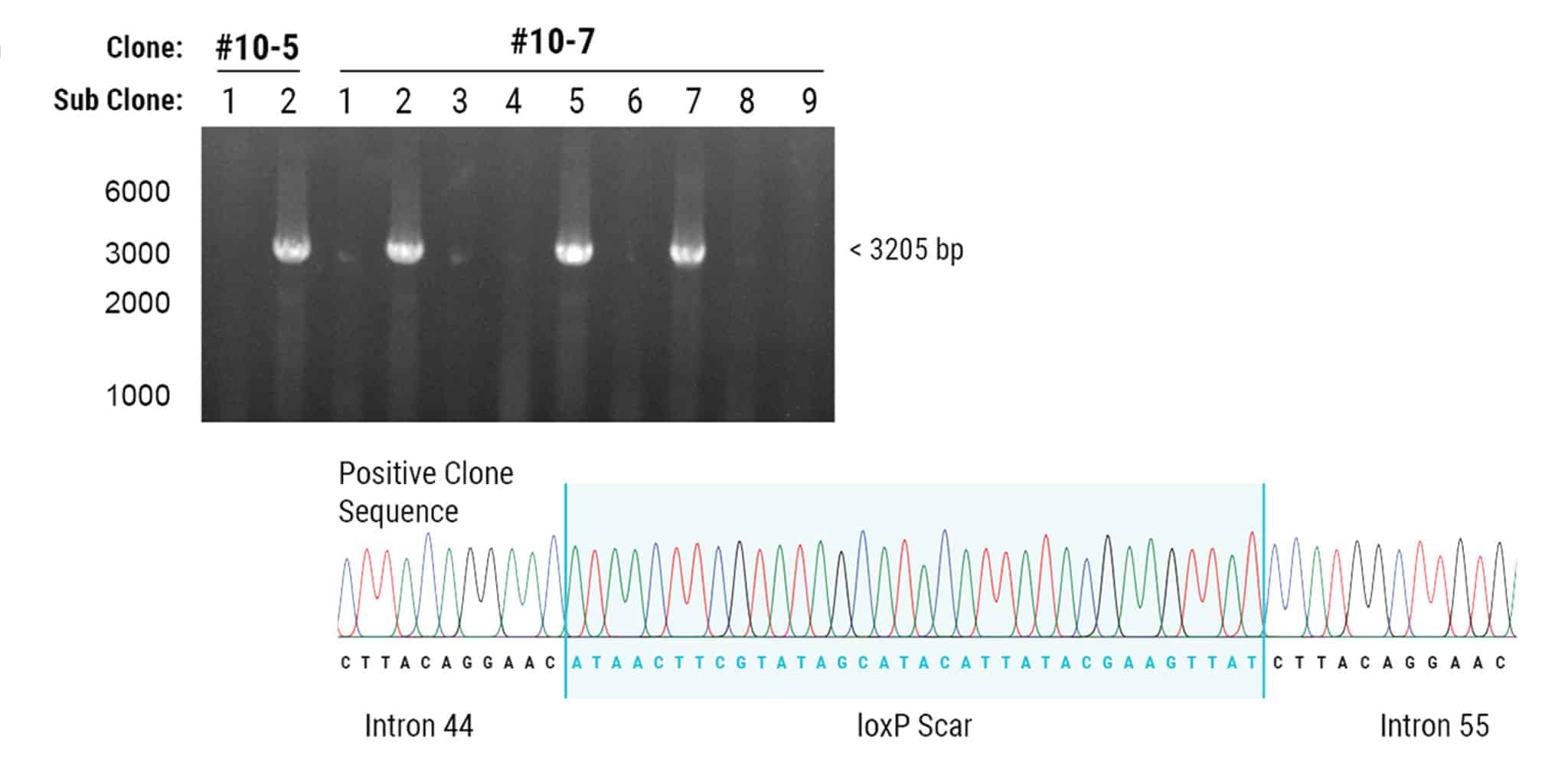
Cre recombinase was transfected into two hiPSC clones containing both loxP sites. Each subclone was PCR genotyped and 4 of 11 revealed a large deletion of 342 kb. Deletion of exons 45-55 was demonstrated in both clones by amplification of loxP scar sites and Sanger sequencing.
Summary
- ssODNs were electroporated into hiPSCs and were integrated without the need for antibiotic selection.
- Sequential delivery of ssODN loxP sites enables editing flexibility as this approach circumvents the requirement for large DNA donor templates.
- MaxCyte electroporation of small ssODNs enabled highly efficient knockin of loxP sites.
- MaxCyte can transfect difficult-to-engineer cells such hiPSCs.
References
- Nakamura A, Shiba N, Miyazaki D, et al. Comparison of the phenotypes of patients harboring in-frame deletions starting at exon 45 in the Duchenne muscular dystrophy gene indicates potential for the development of exon skipping therapy. J Hum Genet. 2017;62(4):459- 463. doi:10.1038/jhg.2016.152
- Kagita A, Lung MSY, Xu H, et al. Efficient ssODN-Mediated Targeting by Avoiding Cellular Inhibitory RNAs through Precomplexed CRISPR-Cas9/sgRNA Ribonucleoprotein. Stem Cell Reports. 2021;16(4):985-996. doi:10.1016/j.stemcr.2021.02.013





