Application Note
Enhancing HIV-1 Vaccine Efficacy and Manufacturing Through CHO Cell CRISPR Engineering
Background
The RV144 HIV vaccine trial, the first trial to demonstrate that vaccination could confer HIV protection, reported a modest 31% efficacy. gp120, a major component of the RV144 trial, was produced in Chinese Hamster Ovary (CHO) cells and lacked N-linked glycosylation sites critical for binding of anti-HIV broadly neutralizing antibodies (bN-mAbs). Researchers believe, at least in part, that the lack of N-linked glycosylation may play a role in the poor efficacy.
Many epitopes bound by bN-mAbs contain oligomannose terminal glycans — early intermediates in the N-linked glycosylation pathway.1 Dr. Berman’s group at University of California Santa Cruz demonstrated using MaxCyte® electroporation that recombinant gp120 (rgp120) production using a HEK 293 cell line deficient in the N-acetylglucosaminyltransferase I enzyme (GnTI- 293; ATCC CRL-3022) resulted in HIV epitopes containing mainly mannose-5 terminal glycans.2 The presence of these glycan moieties improved binding of three major bN-mAb families suggesting use of these rgp120 during vaccination may result in improved efficacy. GnTI- 293 cells, however, are not suitable for large, clinical-scale bioproduction hindering migration to in vivo efficacy studies.
CHO cells continue to be the gold standard for biotherapeutic development, however, the complexity and heterogeneity of protein glycosylation can pose manufacturing and/or immunogenicity challenges. Creation of a CHO cell line deficient in alpha-1,3-mannosyl-glycoprotein 2-beta-N-acetylglucosaminyltransferase (MGAT1) would produce proteins primarily containing oligomannose glycans and could greatly improve HIV gp120 production, purification and potentially vaccine efficacy.
Aim
Generate a CHO MGAT1- cell line via CRISPR gene knockout that maintains growth properties suitable for use in protein manufacturing. In addition, transiently produce rgp120 using the engineered CHO MGAT1- cell line and characterize protein glycosylation patterns and bN-mAb binding.
CHO Electroporation
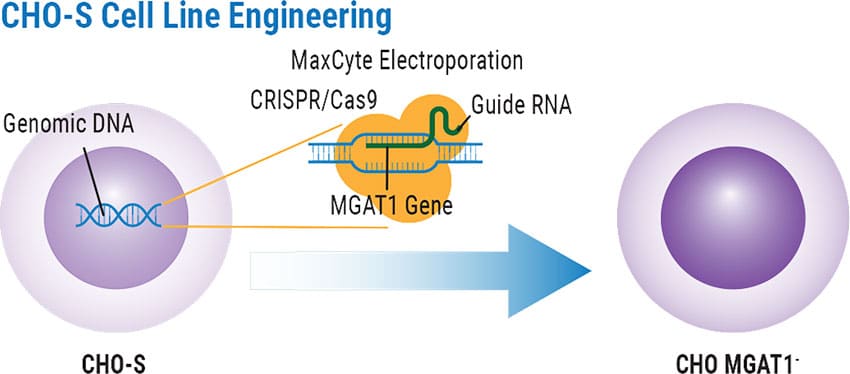
- CHO-S or CHO MGAT1- cells were resuspended in MaxCyte Electroporation Buffer at 2 x 108 cells/mL.
- For CHO MGAT1- construction, CRISPR-Cas9 exonuclease with guide sequence plasmid was added to resuspended CHO-S at a final concentration of 300 μg/mL.
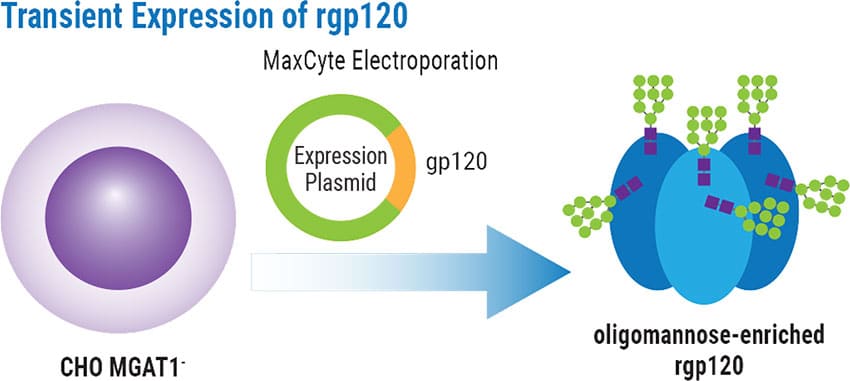
- For transient expression of HIV gp120, an expression plasmid encoding the gp120 from the A244 HIV strain were added to CHO MGAT1- cells.
- Post electroporation, cells were seeded at 4 x 106 cells/mL in OptiCHO media and cultured in 125mL Erlenmeyer shake flasks.
- For gp120 production, cultures were supplemented with 1 mM sodium butyrate 24 hours post electroporation and the temperature lowered to 32°C.
Full methods for CHO MGAT1- screening and selection, as well as glycosylation
analysis and binding assays are detailed in PLoS Biol, 16(8): e2005817, 2018.
Results
CRISPR Delivery & Creation of CHO MGAT1- Cell Line
CHO-S cells were electroporated with a plasmid encoding CRISPR-Cas9, tracrRNA, and a complete guide gRNA targeting the MGAT1 gene. Clones were initially screened for staining with fluorescein-labeled Galanthus nivalis lectin (GNA), a lectin that recognizes glycans with terminal mannose but not complex, sialic acid-containing glycans. As anticipated, the parental CHO-S cell line did not bind GNA. The four GNA-binding clones with the fastest growth rates were transiently transfected with a plasmid encoding A244-rgp120. Secreted rgp120 was purified and analyzed for overall titer and binding to the glycan-dependent bN-mAb PG9.
The selected CHO MGAT1- cell line had rgp120 yields comparable to the parental CHO-S line, as well as similar doubling times and the ability to grow at high density. These characteristics were far superior to the yield, growth time and cell density restrictions of the GnTI- HEK 293 cell line used in previous studies.3 The selected clone was sequenced to confirm disruption of the MGAT1 gene.
Identification of MGAT1- CHO Cells via GNA Binding of Oligomannose
Transient Expression of rgp120
A224-rgp120 transiently produced using the selected MGAT1- CHO cell line or parental CHO-S cells was characterized by MALDI-TOF mass spectroscopy to detail glycan composition. MGAT1-deficiency resulted in rgp120 containing >99% oligomannose glycans compared to only 25% when produced in CHO-S cells.
The homogeneity of glycosylation resulting from MGAT1-deficiency also positively impacted purification. The rgp120 in the RV144 HIV clinical trial showed extensive net charge heterogeneity that necessitated the use of immuno-affinity chromatography for purification greatly reducing yield and increasing manufacturing complexity. rgp120 produced by MGAT1-deficient cells was secreted at high titers with homogeneous glycosylation allowing for purification using more conventional chromatography.
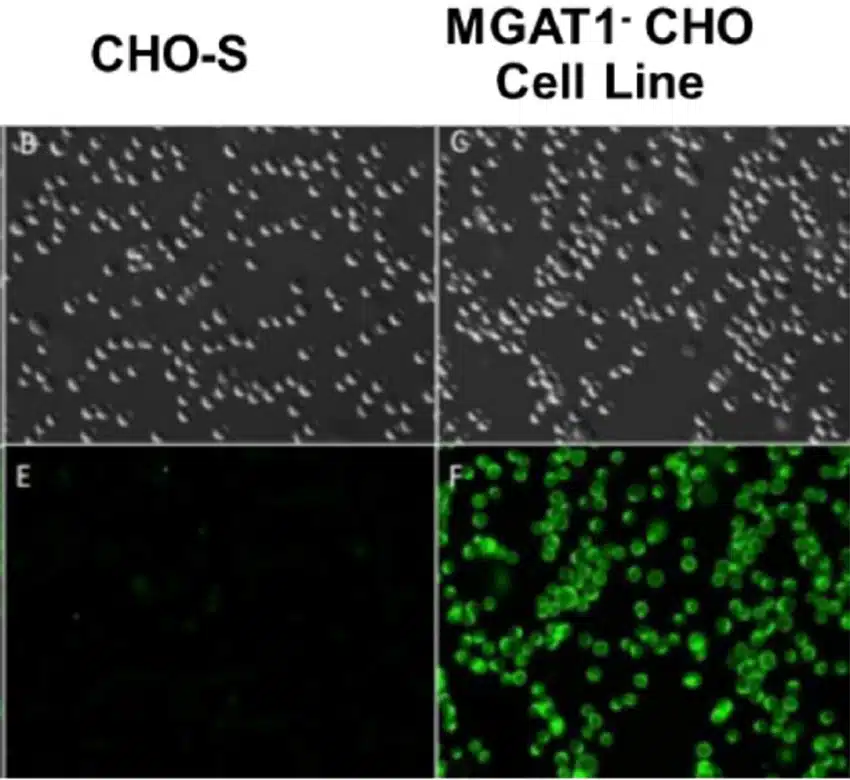
Figure 1. CHO-S parental cells or those electroporated with MGAT-targeting CRISPR machinery were incubated with fluorescein labeled GNA to identify cells with surface proteins containing terminal mannose residues.
MGAT1-Deficient CHO Cells Produce rgp120 Containing >99% Oligomannose Glycans

Figure 2. Purified rgp120 from the selected CHO MGAT1- cell line or the parental CHO-S cell line were analyzed via MALDI-TOF mass spectroscopy.
MGAT1-Deficiency Produces rgp120 with Improved Binding to Multiple bN-mAbs
Binding of rgp120 produced in CHO-S or MGAT1- CHO cells to a panel of bN-mAbs was assessed via fluorescence immunoassay.3 All but a single antibody had modest to significant improvements in binding to rgp120 produced in the MGAT-deficient cells. These results suggest that altering glycosylation while leaving the amino acid sequence unchanged can positively impact bN-mAb binding. Examination of the effects of oligomannose engineering on in vivo immunogenicity and vaccine efficacy are critical next steps.
Improved Binding of V1/V2 and V3 Domain bN-mAbs to rgp120 Produced in MGAT1-deficient CHO Cells
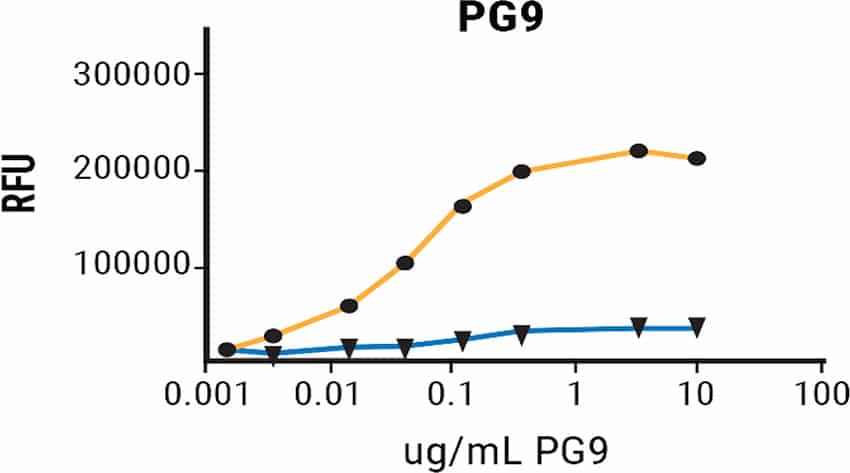
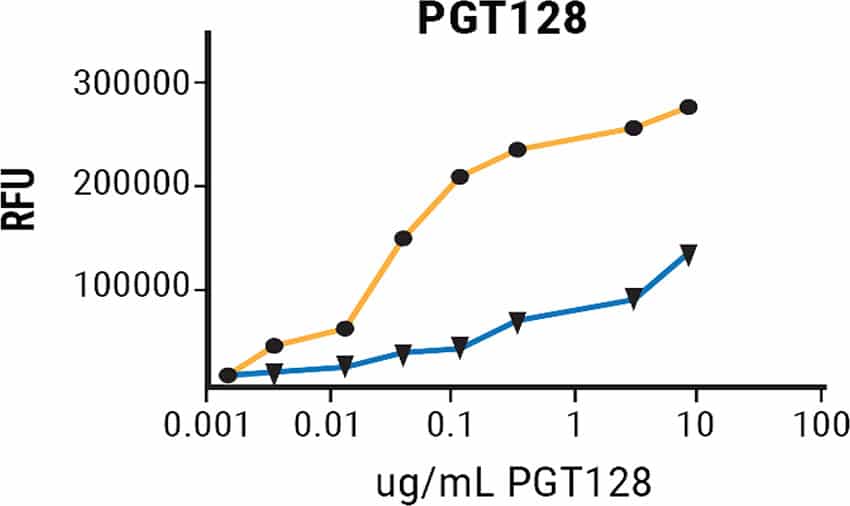

Figure 3. A224-rgp120 purified from CHO-S or MGAT1- CHO cells were coated onto microtiter plates and binding of a panel antibodies with broadly neutralizing capacity examined via FIA.
Conclusion
MaxCyte’s high performance cell engineering technology delivered the CRISPR-Cas9 machinery that resulted in levels of efficiency and CHO-S cell viability that enabled the rapid generation of an MGAT1-deficient CHO cell line. The established MGAT1- CHO cell line maintained robust growth in serum-free medium, was able to grow at high cell densities in suspension and produced rgp120 at high titers upon transient transfection. rgp120 produced using the newly engineered MGAT1- CHO cells gave rise to the desired early oligomannose glycans which in turn enhanced the binding of key bN-mAbs. This cell line promises to be a critical tool in the clinical translation of more efficacious HIV vaccines.
References
- High-mannose Glycan-dependent Epitopes Are Fequently Targeted in Boad Neutralizing Antibody Responses during HIV-1 Infection. (2012) J Virol, 86(4):2153-2164.
- Glycan Modification to the gp120 Immunogens Used in the RV144 Vaccine Trial Improve Binding to Broadly Neutralizing Antibodies. (2018) PLoS ONE, 13(4):e0196370.
- CRISPR/Cas9 Gene Editing for the Creation of an MGAT1-deficienct CHO Cell Line to Control HIV-1 Vaccine Glycosylation. (2018) PLoS Biol, 16(8): e2005817.





