Scientific Brief
A Fully Optimized CRISPR Workflow for Drug Discovery in T Cells
Abstract
Immune-mediated inflammatory diseases (IMIDs) are characterized by excessive tissue inflammation, uncontrolled antibody production and an imbalance of immunoregulatory cytokines. Drugs that enhance production of anti-inflammatory cytokines could provide therapeutic benefit for patients with IMID and other autoimmune disorders. Here we present a pilot study for the end-to-end, whole-genome CRISPR knockout screening of primary CD4+ T cells to discover new therapeutic targets. High-efficiency MaxCyte® electroporation of Cas9 protein and optimized cytokine-positive cell enrichment were combined with the SLICE (sgRNA lentiviral infection with Cas9 protein electroporation) screening protocol. This framework reproduced validated hits and was used to identify new genetic targets with a view to drug development.
Background
Lentiviral CRISPR screening in primary T cells presents significant challenges due to the low transduction efficiency of lenti-Cas9 vectors and short T cell lifespan ex vivo. The SLICE protocol was previously described as a solution for whole-genome screens in human T cells with a combined approach of lentiviral gRNA library transduction followed by electroporation of Cas9 protein. Here, SLICE screening was adapted for use with CD4+ T cells with MaxCyte electroporation and optimized cytokine assays and transduction methods from GSK®. The MaxCyte ExPERTTM platform provided scalability up to 3 x 108 cells and high-efficiency gene editing with low cell toxicity, all critical parameters for handling sensitive primary T cells in a genome-wide CRISPR screen.

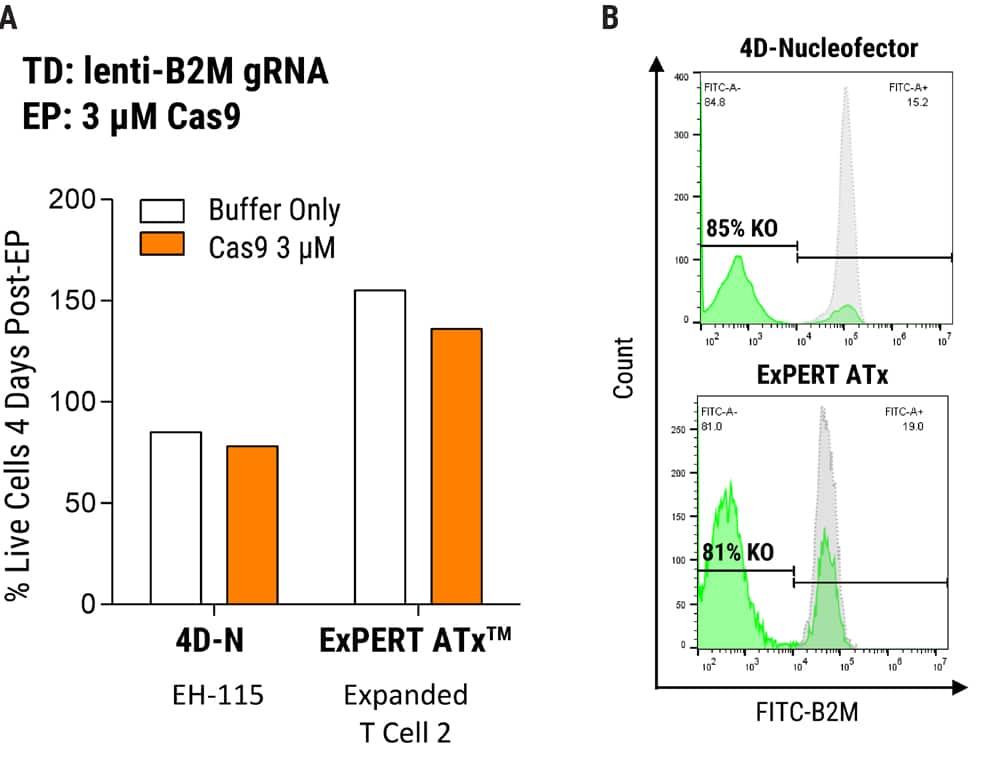
Figure 1. MaxCyte Electroporation Provides High Knockout Efficiency with Superior Cell Recovery A) Following the delivery of a single gRNA with an optimized transduction protocol, MaxCyte ExPERTTM electroporation (EP) of Alt-R® SpCas9 (IDT) led to >80% knockout efficiency and 2x better cell recovery in a single round of transfection 4 days post EP. Conversely, low throughput capabilities and cell toxicity were observed using the Lonza 4D-Nucleofector®, which resulted in 50% lower cell recovery 4 days post EP. B) MaxCyte delivered >80% knockout, as shown by FACS (Fluorescence-activated cell sorting) analysis of a representative gene target, beta-2-microglobulin (B2M, FITC-label.)
Figure 2. Optimization of MaxCyte EP in SLICE Transduced T Cells. 1 x 107 cells were transduced with gRNA targeting B2M followed by electroporation with either GFP mRNA or Cas9 protein using the ExPERT ATx® and OC100 x 2 processing assembly, 100 µl volume, program Expanded T cell 2. A) Uptake of mRNA was evaluated by FACS analysis of GFP expression, yielding 93% and 86% GFP-positive cells at 24 and 48 hours post EP, respectively. B) B2M knockout was assayed by FACS (1 µM or 3 µM Cas9 protein concentration) and resulted in 81% protein depletion under both conditions, 4 days post EP.
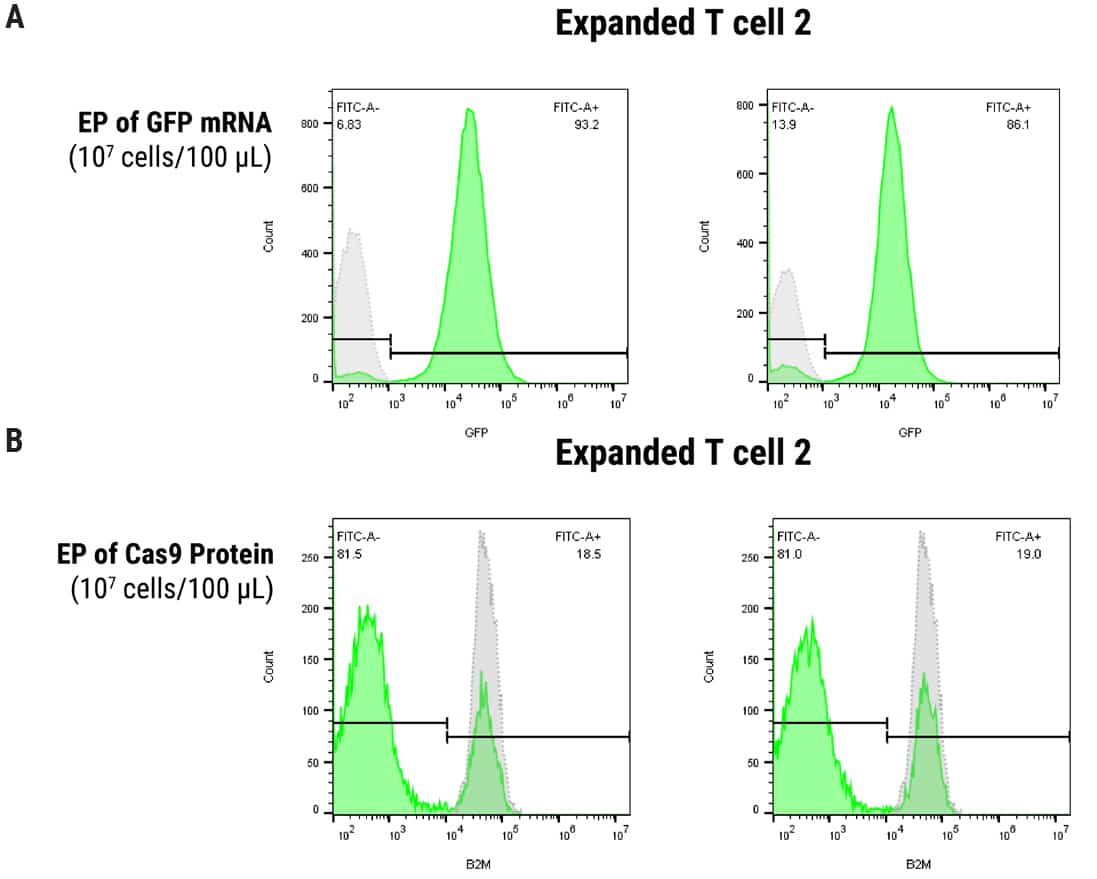
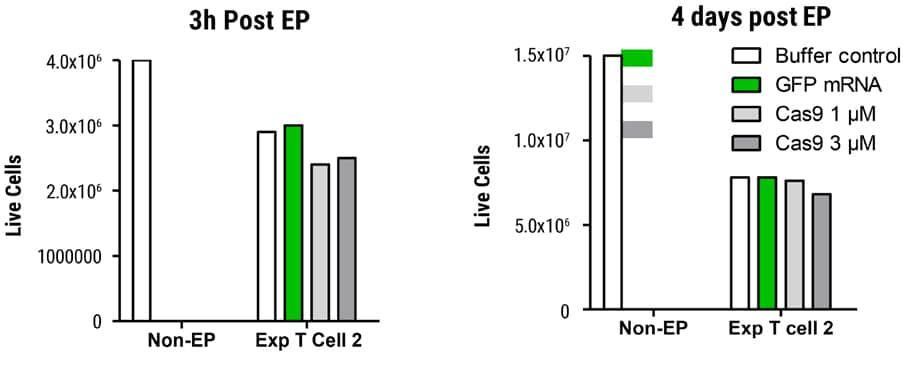
Figure 3. Viability in SLICE Transduced T Cells after MaxCyte EP Transduced cells were electroporated with GFP mRNA or Cas9 protein and cell viability determined 3 hours and 4 days post EP. Cells were counted on the Nucleocounter® NC-200TM (acridine orange DAPI). Live cell recovery was essentially equivalent between GFP mRNA or Cas9 protein-electroporated samples on day 4 post EP.
Figure 4. Electroporation Scale-Up and Maintenance of T Cell Phenotype. To model the large-scale cell handling requirements of a genome-wide screen, the ExPERT ATx® with CL 1.1 processing assembly was used to electroporate 3 x 108 T cells with Cas9 protein. Cells were pre-transduced with B2M lenti-gRNA; Cas9 protein was delivered at 3µM concentration in a 3 mL volume using the program Expanded T Cell 2. A) B2M protein depletion was 82% on day 4 post EP. B) Live cell recovery was high, assayed at 3h and 4 days post EP. Cas9 electroporated cells exhibited lower recovery, as expected, but in sufficient quantity for downstream experiments. C) Cells were monitored throughout the workflow for markers of T cell immunophenotype. A representative donor exhibits stable expression of multiple genes throughout T cell stimulation, gRNA transduction and Cas9 electroporation.

Analysis of gRNA Enrichment
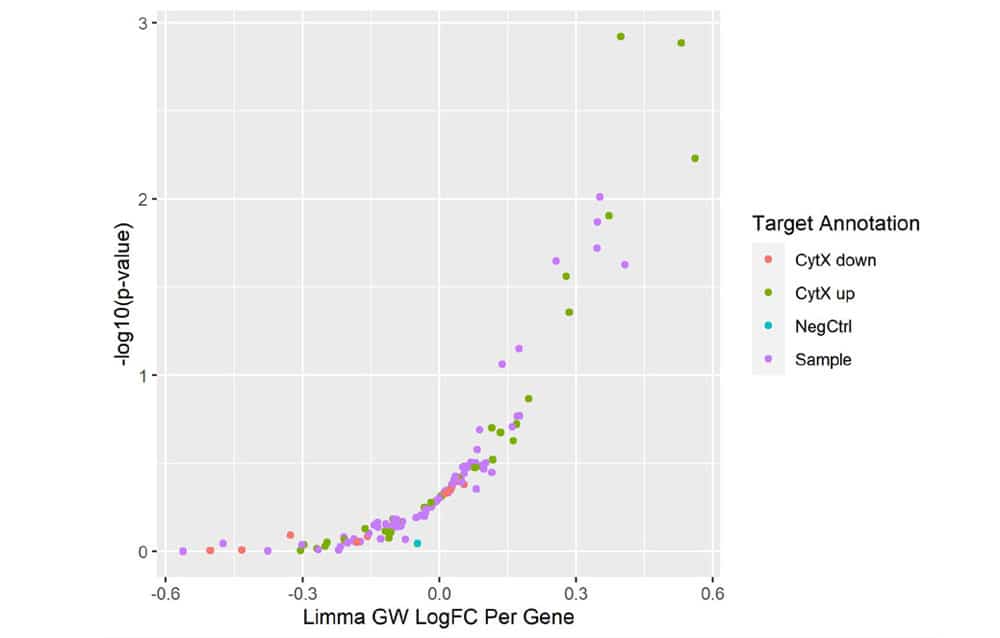
Figure 5. Mini-Screen Results. A focused screen was performed on T cells from two donors according to the modified SLICE workflow shown above with 1000 gRNAs, 4 gRNAs/gene, 2 x 106 cells. Following FACS enrichment for both transduced and cytokine expressing cells (GSK, data not shown), cell populations were analyzed by Next Generation Sequencing (NGS) using the MiSeq® system (Illumina, Inc.) Deconvolution of NGS data clearly identified 4 top-ranked genes which were known to strongly increase cytokine X (CytXUp), thus validating the performance and sensitivity of this screening workflow.
Summary
- The SLICE protocol with cytokine assays were fully optimized in a mini-screen approach.
- T cell characteristics were verified and maintained by immunophenotyping at multiple steps during the screen.
- MaxCyte electroporation enables optimal editing efficiency and scalability to 3 x 108 cells in a single tranfection for whole-genome primary T cell screens.
- MaxCyte electroporation ensures high cell viability for sensitive cell populations.
- Potential future experiments:
- Genome-wide CRISPR screen for targets that regulate cytokine expression
- CRISPR RNP-based validation of individual hits
- In-depth interrogation of target druggability, gene expression, gene pathways and toxicity
- Potential application to a wide range of future CRISPR screens in primary T cells
References
- Webinar: Development of a genome-wide CRISPR screen in CD4+ T cells to identify drug targets for immune-mediated inflammatory diseases
- Shifrut E, Carnevale J, Tobin V, et al. Genome-wide CRISPR Screens in Primary Human T Cells Reveal Key Regulators of Immune Function. Cell. 2018;175(7):1958-1971.e15. doi:10.1016/j.cell.2018.10.024





