Application Note
A Novel Cell-Based Assay for Ubiquitin Drug Discovery
Background
Ubiquitin (Ub) is an abundant regulatory polypeptide involved in most cellular processes, added to proteins through a process known as ubiquitylation. The dysregulation or subversion of ubiquitin pathways has been implicated in conditions including viral infection, hereditary neurodegenerative diseases and cancer. Unfortunately, the complexity of the system and a shortage of high-throughput screening assays have caused ubiquitin drug discovery to lag behind many other target classes, such as kinases, GPCRs or ion channels. Ubiquitylation is an enzymatic cascade of activation (E1), conjugation (E2) and ligation (E3). Substrate specificity is conferred by the E3 ligases, making this class of over 600 putative enzymes a potential therapeutic target. Although several cell-free assays to detect E3 ligase inhibitors have been developed using engineered tandem ubiquitin-binding entities (TUBEs) that bind polyubiquitin chains, these are labor intensive, not readily adaptable to new targets, and lack biological context. Developing a cell-based assay for high-throughput screening in physiological conditions would address these issues and be a step forward in E3 ligase drug discovery. Cell-based assay development typically relies on stable cell lines, which can be challenging to produce, requiring multiple rounds of transfection, selection and screening; a new cell line is needed for each new target. MaxCyte® Electroporation® enables the rapid preparation of transiently transfected, Assay Ready Cell banks, simplifying cell-based assay development and providing greater target flexibility.

Aim
This study describes the development of a novel high-throughput, cell-based Ubiquitin Ligase Profiling assay (ULP)1. MaxCyte’s Electroporation enabled three plasmid co-transfection of HEK293 cells to generate Assay Ready Cell banks for method validation and compound screening without the need for stable cell line development.
Methods
Ubiquitin Ligase Profiling Method
- 1 mL aliquot of Assay Ready Cells was rapidly defrosted (37°C), then washed and resuspended at 1x106 cells/mL in warm assay media
- 10 µL cells per well were dispensed into 384-well assay ready compound plates
- Assay plates were incubated for 20 hours in a tissue culture incubator
- Cells were brought to room temperature, and luciferase assay reagents were added
- Luminescence was read after 30 minutes incubation in the dark
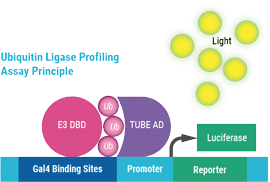
Under normal physiological conditions, the E3 ligase-DNA Binding Domain fusion (E3 DBD) poly-autoubiquitylates and binds to Gal4 sites on the reporter plasmid. The TUBE domain of the TUBE-Activation Domain (TUBE AD) binds the polyubiquitin chain, bringing the activation domain closer to the promoter, resulting in increased transcription of firefly luciferase, measured by luminescence. In the presence of an E3 ligase inhibitor, autoubiquitylation is inhibited; there is no recruitment of the activation domain, thus only basal transcription of firefly luciferase.
Results
The Ubiquitin Ligase Profiling (ULP) assay detects physiological E3 ligase activity.
The ULP method was validated by detecting autoubiquitylation of three E3 ligases; Rnf8, Chfr and Traf6. Firefly luciferase signal was detected only when a functional E3 ligase was included. Conversley, only basal signal was detected when a catalytically dead E3 mutant or empty assay plasmids were used (Figure 1). These results confirmed that the ULP method detects E3 ligase autoubiquitylation in a cellular context.
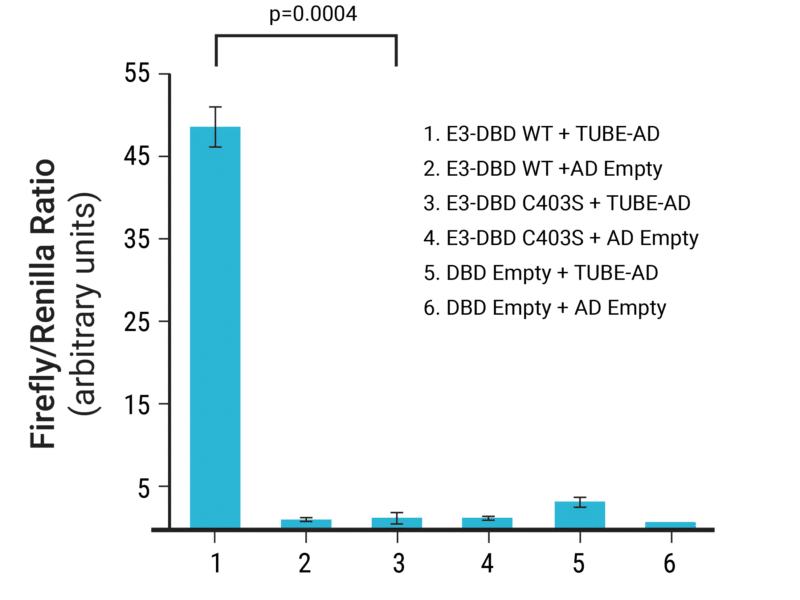
Figure 1. Small-scale proof-of-concept using chemical transfection demonstrates assay specificity. HEK293 cells were transfected with combinations of Rnf8 assay vectors and empty control plasmids (n=3). The induction of firefly luciferase expression was normalized to Renilla luciferase expression to account for variations in transfection efficiency with chemical transfection. Functional E3 ligase = WT, catalytically-dead E3 ligase = C403S.
Cryopreserved Assay Ready Cells are a robust platform for high-throughput screening with the ULP assay.
MaxCyte Flow Electroporation was used to generate Assay Ready Rnf8, Chfr or Traf6 cells, which were aliquoted and cryopreserved. The ULP method was miniaturized to a 384-well format for high-throughput screening using cryopreserved, banked cells. The assay was validated by screening a compound test set and met industry standards1 (Figure 2).
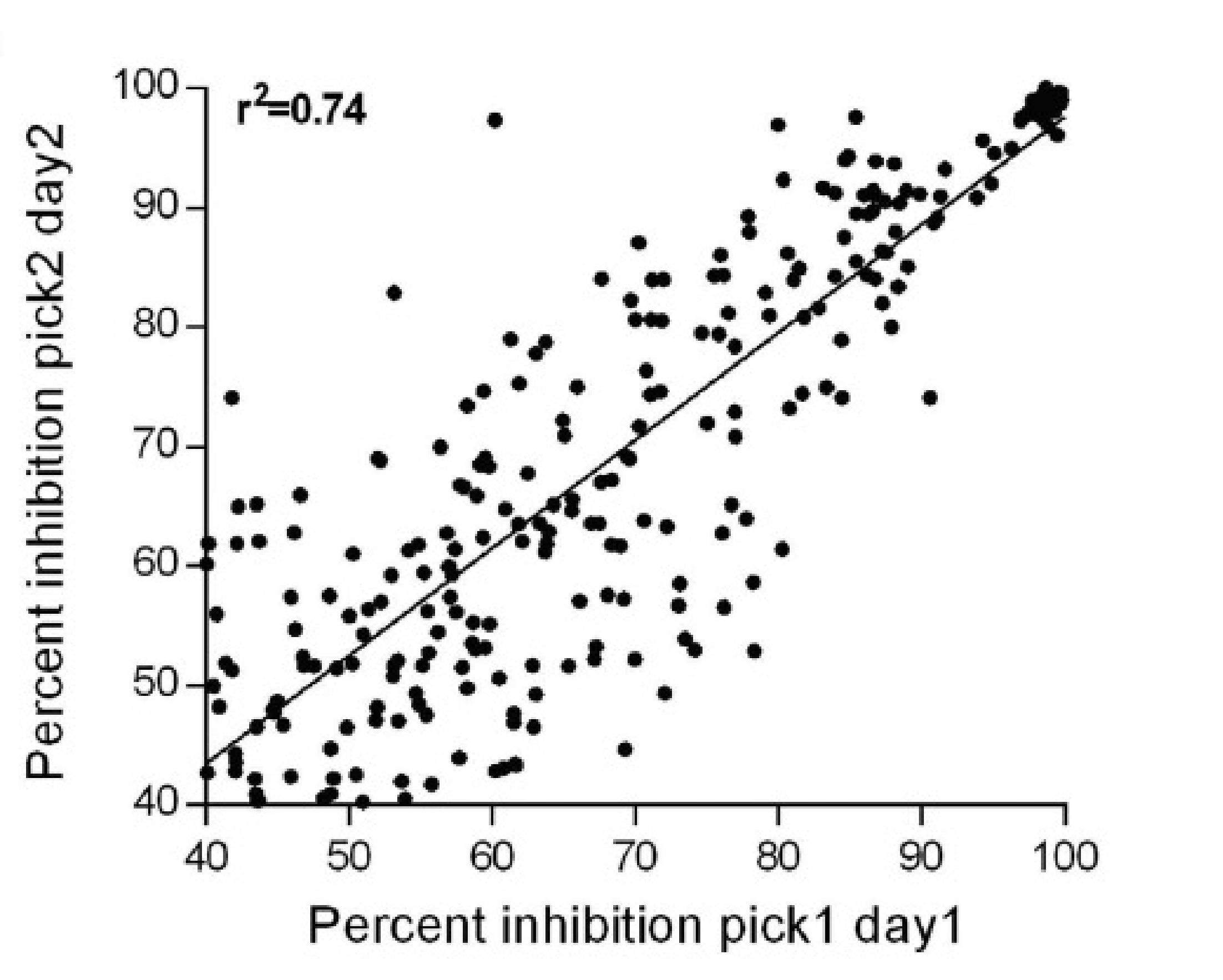
Figure 2. Test set screening for ULP assay validation. 9,846 randomly plated test compounds were screened on two days. Firefly luciferase activity was measured, and the percentage inhibition for compounds with ≥40% inhibition on both days was plotted. The Rnf8 ULP assay Z’ factor (0.68 +/– 0.02 SEM) and Coefficient of Variation (8.6% +/– 1.1 SEM) were determined.
Primary screening identified a subset of compounds that selectively inhibited Rnf8 E3 ligase activity.
Rnf8 is involved in the cellular DNA damage response, making it an exciting target for therapeutic intervention.
A primary screen in Rnf8 electroporated cells identified 3,327 hits (≥ 40% signal inhibition), from which known frequent hitters were eliminated. Compound IC50 was determined by secondary screening in MaxCyte electroporated, Assay Ready Cells. A comparison of compound IC50 against the three E3 ligases (Figure 3) was used to identify 127 hits with Rnf8 selectivity. These results validated using Assay Ready Cells in the high-throughput ULP assay to identify inhibitors of a specified E3 ligase. Following biochemical analysis of the hits (data not shown), compound-1 was chosen for further study.
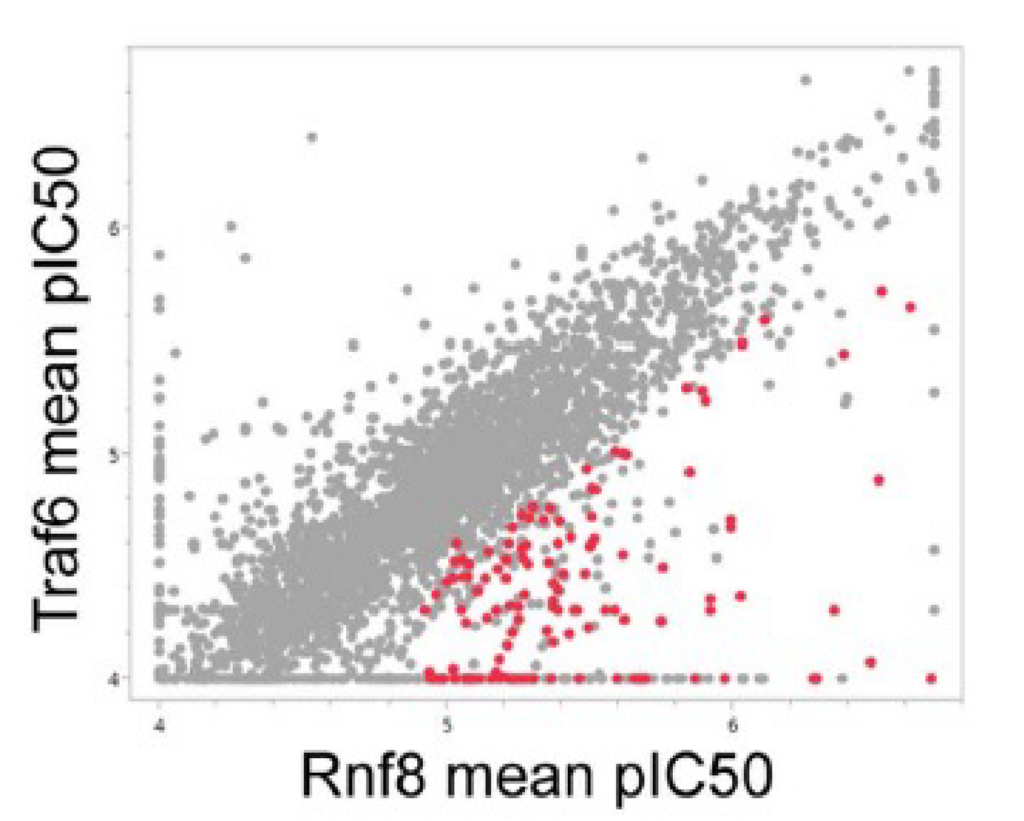
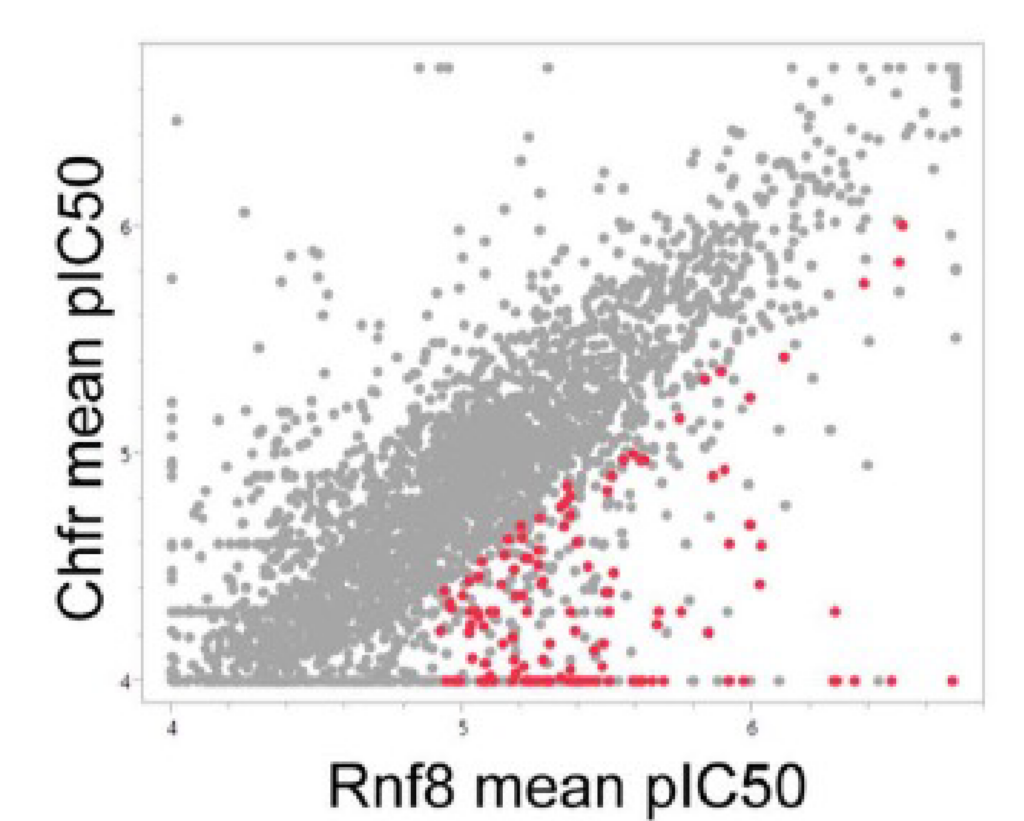
Figure 3. ULP assay screening identified Rnf8-specific inhibitors. The ULP assay was used to determine compound IC50 against Rnf8, Traf6 and Chfr. Mean pIC50 was compared between (a) Rnf8 and Traf6 and (b) Rnf8 and Chfr. Screening identified 127 compounds with Rnf8 pIC50 ≥4.9 and ≥0.5 log unit selectivity for Rnf8 versus both Traf6 and Chfr (shown as red dots).
Compound-1 blocks Rnf8 activity in DNA damage response.
In response to DNA damage, several factors are recruited to the site of DNA double-stranded breaks, forming repair foci. ATM kinase activity recruits and phosphorylates Mdc1, then Rnf8 binds phosphorylated Mdc1, recruiting 53bp1 through a ubiquitylation-dependent mechanism.
The effect of pre-treatment with compound 1 on the recruitment of Mdc1 (upstream of Rnf8) and 53bp1 (downstream of Rnf8) to DNA damage repair foci was compared to pre-treatment with either DMSO or an ATM kinase inhibitor. As expected, the ATM inhibitor blocked the recruitment of Mdc1 and 53bp1, while compound 1 inhibited only the recruitment of 53bp1 (Figure 4).
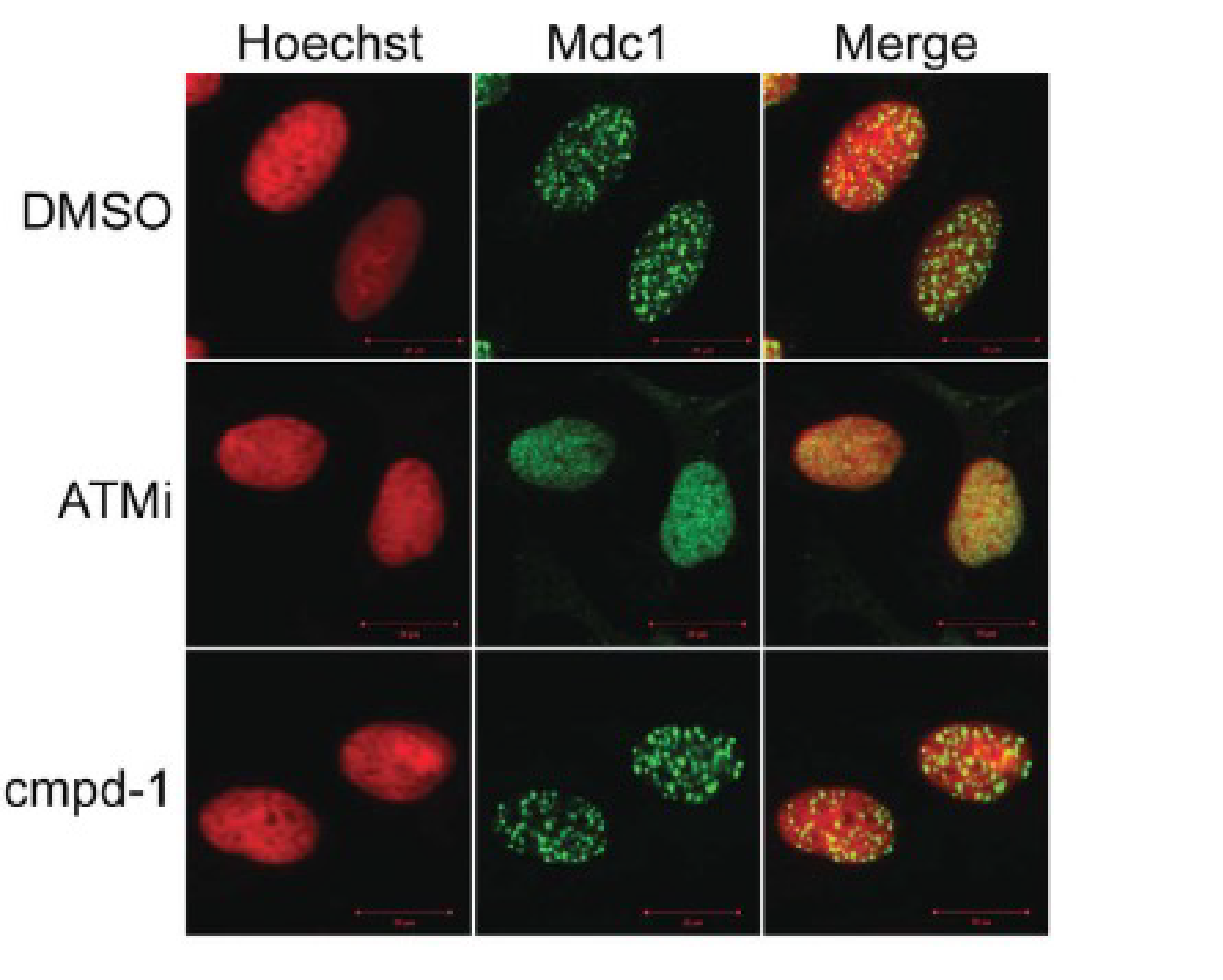
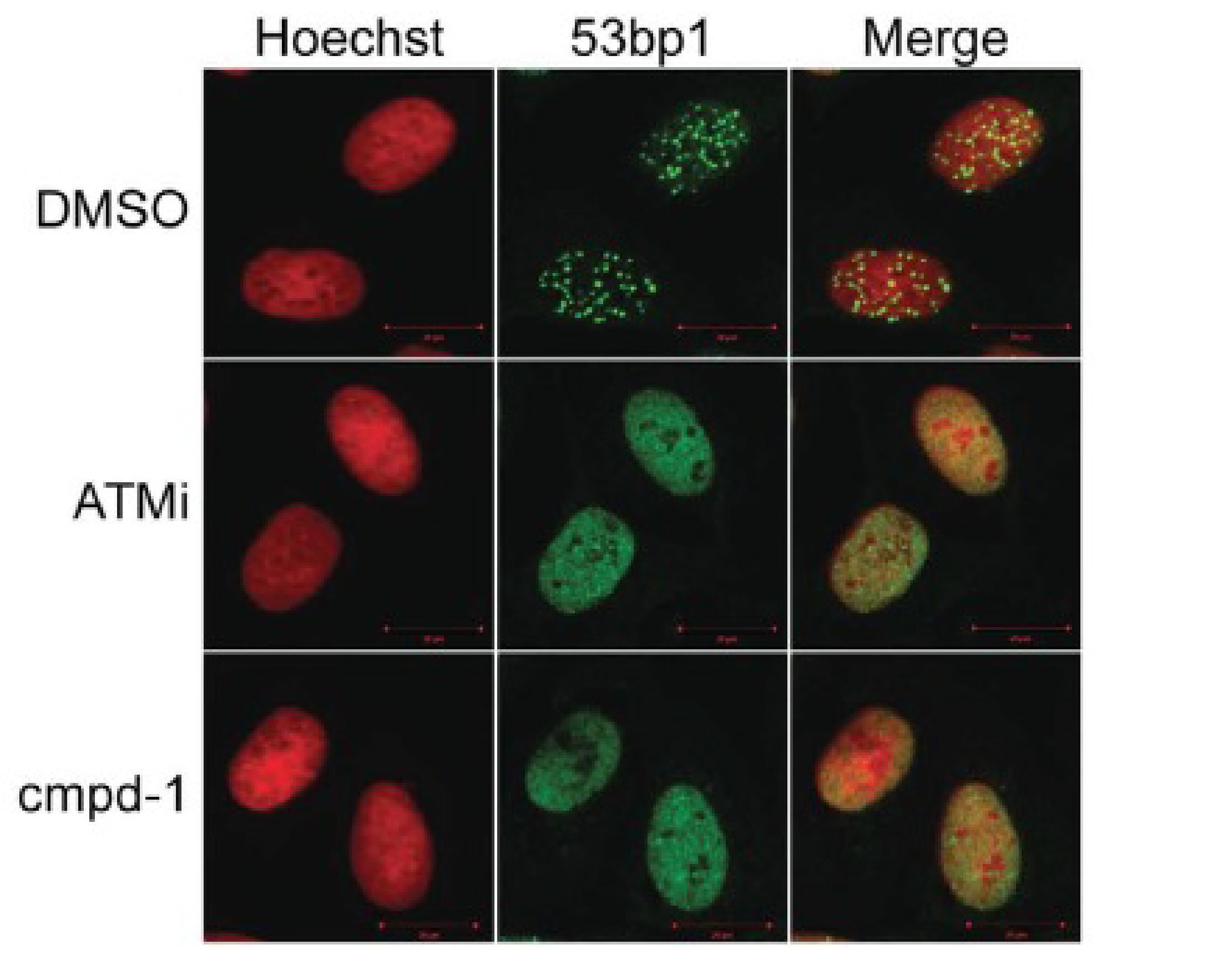
Figure 4. Compound-1 inhibits Rnf8 signaling in DNA damage response. U2-OS cells were treated with DMSO, ATM inhibitor (ATMi) at 12.5 µM or compound-1 (cmpd-1) at 50 µM for 3 hours followed by irradiation with 10Gγ. Cells were imaged by confocal microscopy with either (a) anti-Mdc1 or (b) anti-53bp1 followed by Hoescht staining.
Conclusion and Future Applications
The MaxCyte STX™ delivered multiple plasmids simultaneously to large numbers of cells to prepare transiently-transfected, Assay Ready Cells to streamline ULP method development and validation. Using this novel assay, the researchers screened a small compound library and identified several highly-selective inhibitors of the Rnf8 E3 ligase, from which a potent, biologically-active compound was chosen for further study.
Assay Ready Cells enable method development and high-throughput screening without stable cell line development, saving time and resources. Creating Assay Ready Cell banks of up to 2x1010 cells by a single MaxCyte® electroporation yields assay consistency that would not be achievable when performing a small-scale chemical transfection for each round of screening. MaxCyte electroporation can be used to prepare Assay Ready Cells from virtually any cell type, equipping researchers with the flexibility to readily adapt existing assays to new targets of interest for faster drug discovery.
References
-
Maculins, T. et al. A Generic Platform for Cellular Screening Against Ubiquitin Ligases. Sci. Rep. 6, 18940; doi: 10.1038/srep18940 (2016)
* Since the publication of this study, the new ExPERT STx® instrument has been released to include enhanced software, an improved user interface and an integrated touch screen.





