Scientific Brief
Generation of Heterozygous HLA-C iPSCs by ssODN Electroporation for Allogeneic Transplantation
Abstract
Human-induced pluripotent stem cells (iPSCs) have tremendous potential for regenerative medicine. While there is an ongoing effort to recruit human leukocyte antigen (HLA) homozygous donors for allogeneic transplantation, a significant challenge with this approach is the susceptibility of these cells to host natural killer (NK) cell cytotoxicity. Donor iPSCs that express a single HLA type, such as HLA-C1, may mismatch with NK cells that express two types of killer cell immunoglobulin-like receptors (KIR). This results in NK cell activation and subsequent attack on the donor cells. MaxCyte® enabled the development of HLA heterozygous iPSCs by delivering CRISPR-Cas9 RNPs and single-stranded oligodeoxynucleotides (ssODN) for gene editing. Hotta et al., modified clinical-grade HLA homozygous iPSCs (with two HLA-C alleles) using two ssODN donor templates to generate heterozygous HLA-C1 and HLA-C2 cells. One-step biallelic modification was used to create custom-edited HLA-C iPSCs with allogeneic transplantation potential.

Figure 1. NK Cell-Mediated Cytotoxicity Activated by KIR and HLA Mismatching. KIR 2DL3 and KIR 2DL1 on NK cells will bind HLA-C1 and HLA-C2, respectively. A) If the NK cell interacts with a homozygous HLA-C1 iPSC-derived donor cell, then the lack of HLA-C2 will activate the NK cell and induce a cytotoxic response. B) However, if the iPSC-derived donor cell expresses both HLA-C1 and HLA-C2, then NK cell activity is suppressed.
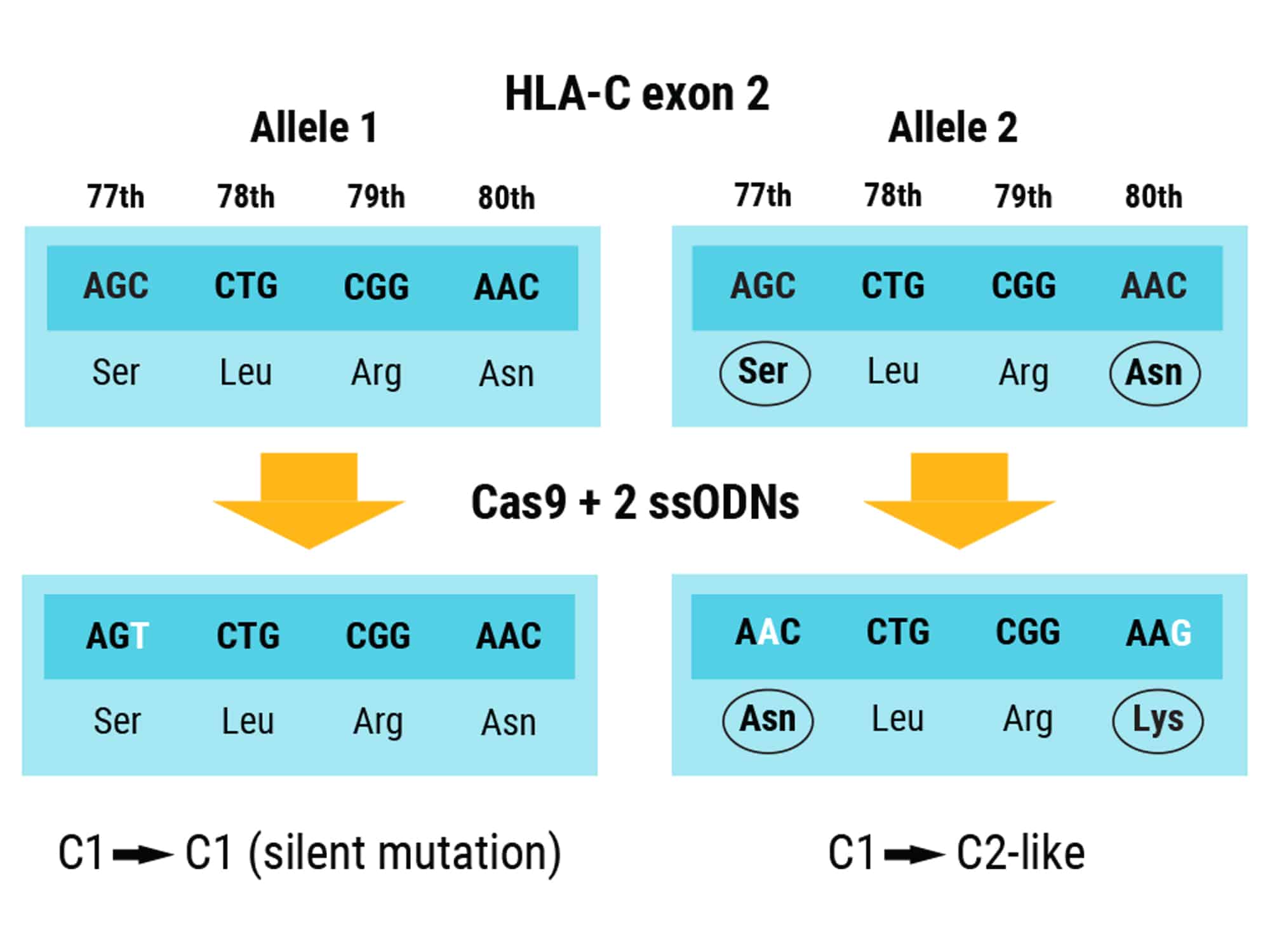
Figure 2. Modification of Both HLA-C1 Alleles Using Multiple ssODNs. CRISPR-Cas9 RNP combined with two ssODN DNA donor templates simultaneously edits both alleles of HLA-C1 in exon 2. One ssODN prevents Cas9 from re-cutting the first HLA-C1 allele by introducing a silent mutation. The other ssODN converts the second allele to HLA-C2 by substituting serine to asparagine at position 77 and asparagine to lysine at positions 77 and 80, respectively.
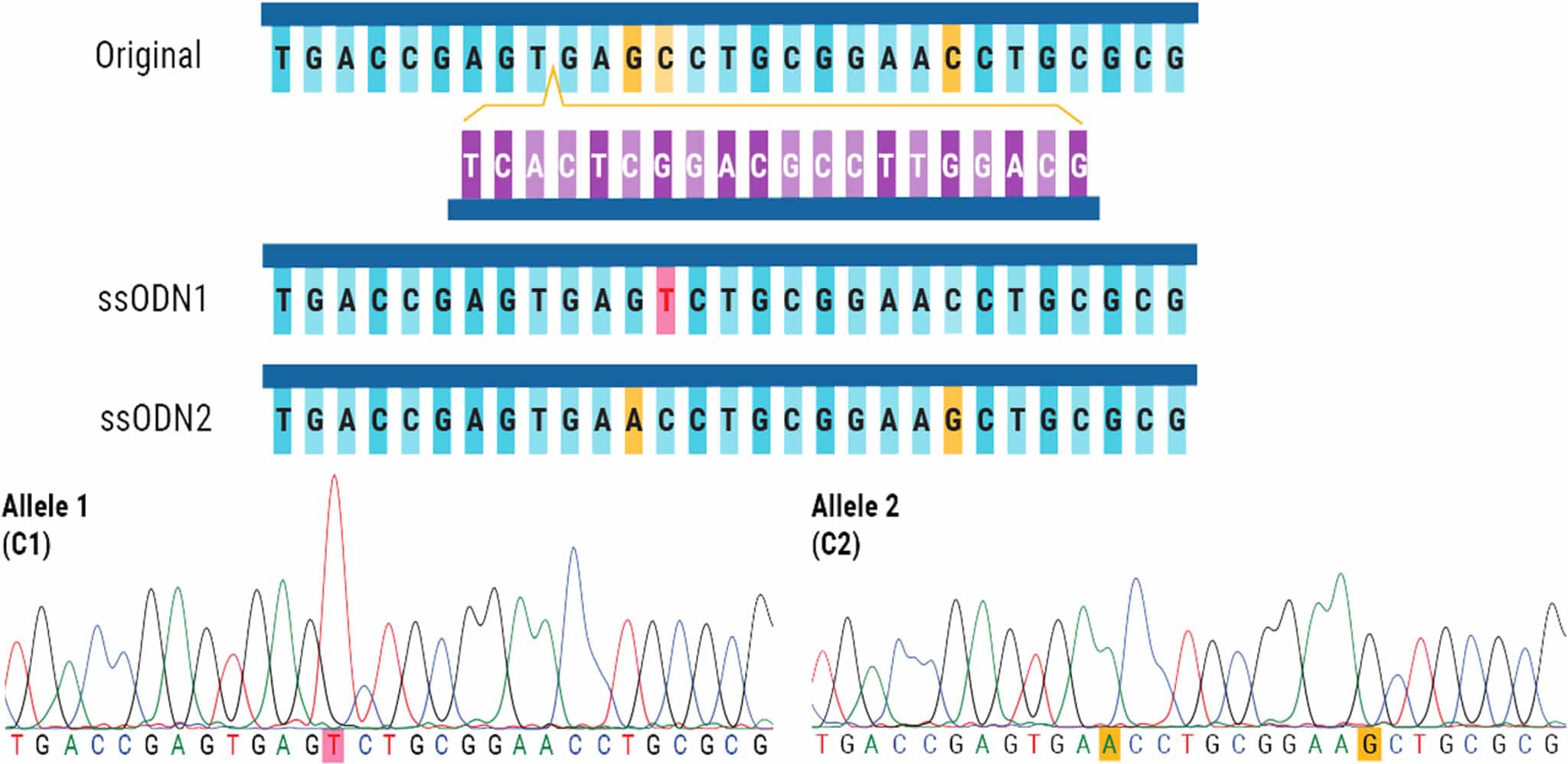
Figure 3. Successful, One-Step Biallelic Editing of iPSCs. The original DNA sequence is shown with the corresponding sgRNA. The cleavage site in the DNA sequence is marked with a yellow arrow. ssODN1 contains a silent mutation highlighted in pink, while mutations in ssODN2 are highlighted in yellow. When analyzing bulk edited cells, each allele was found to have HDR efficiencies of approximately 24–28%. After Sanger sequencing analysis, one out of twenty-nine clones contained the expected modifications in both alleles.
Summary
- HLA heterozygous iPSCs were generated using CRISPR-Cas9 RNPs and ssODNs donor templates.
- MaxCyte® electroporation of multiple ssODNs enabled highly efficient one-step biallelic engineering of iPSCs.
- MaxCyte electroporation can be used to transiently transfect a variety of cell lines such as iPSCs and stem cells with high transfection efficiencies and cell viability.
References
- Kagita A, Lung MSY, Xu H, et al. Efficient ssODN-Mediated Targeting by Avoiding Cellular Inhibitory RNAs through Precomplexed CRISPR-Cas9/sgRNA Ribonucleoprotein. Stem Cell Reports. 2021;16(4):985-996. doi:10.1016/j.stemcr.2021.02.013
This content was adapted from Kagita et al. 2021 under the Creative Commons license
Attribution 4.0 International (CC BY 4.0)





