MaxCyte is excited to attend the 2025 European Society of Gene and Cell Therapy (ESGCT) Congress, taking place in Seville, Spain. We’re proud to be part of this prestigious event and invite you to join us for our luncheon symposium and panel discussion on Thursday, October 9, from 1:30 to 2:30 p.m. This in-person-only session will feature three distinguished speakers and showcase how MaxCyte’s technology is enabling the next generation of cell and gene therapies – from bench to clinic.
This conference is only accessible by attending in person – there are no virtual or recorded components. Attendees will learn about the latest advancements in cell and gene therapy and get to network with peers and industry leaders.
Additionally, MaxCyte is hosting a half-day forum ahead of ESGCT 2025 to explore advances in non-viral cell engineering. Connect with leading researchers, join interactive roundtables and experience hands-on demos with the ExPERT™ platform on Monday, October 6, starting at 1:00 p.m.
Topics include:
- CAR/TCR screening
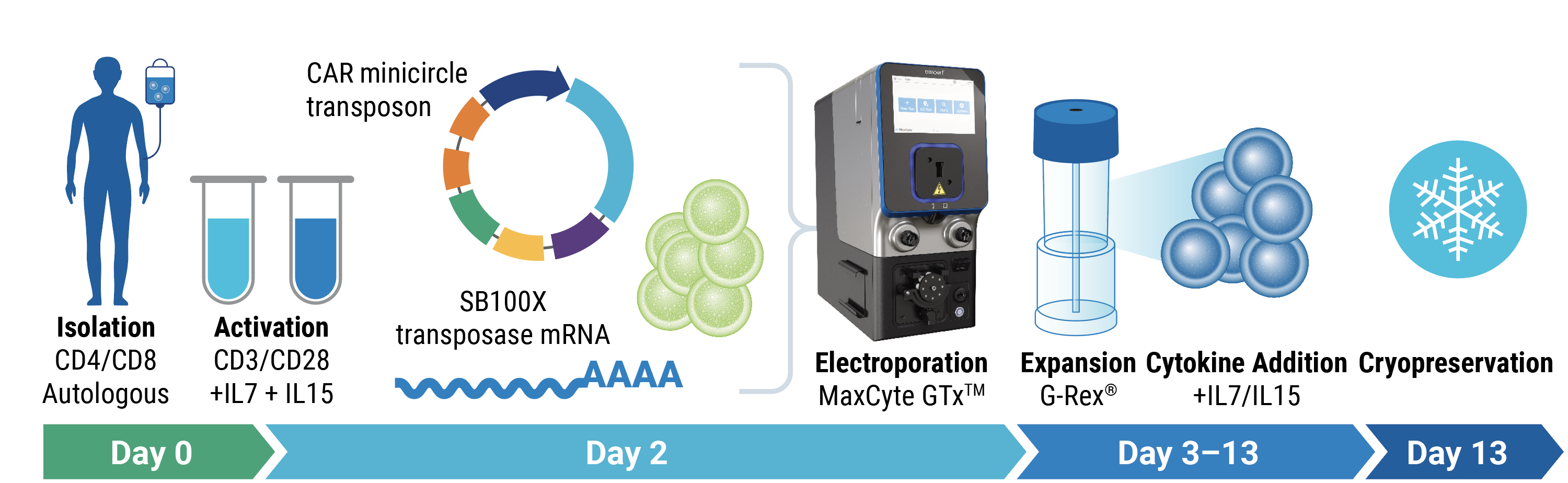
- Transposon engineering
- Gene editing risk assessment
- GMP and scale-up challenges
We hope you can take the time to meet with us in Seville – even just to say "hi" and connect. We look forward to seeing you there!
Come visit MaxCyte at booth #C17-C19 at ESGCT and explore our cGMP-compliant, non-viral cell engineering platform designed to streamline and accelerate the development of cell and gene therapies, from concept to clinic. Discover how our technology empowers seamless, scalable and high-performance cell modification to support your therapeutic pipeline.
Featured presentation
Lunchtime Symposium: Scalable Non-Viral Technologies in Action from Concept to Clinic
Thursday, October 9, 2025, from 1:30 - 2:30 p.m. in Room Parallel D
Presenters



Poster presentations
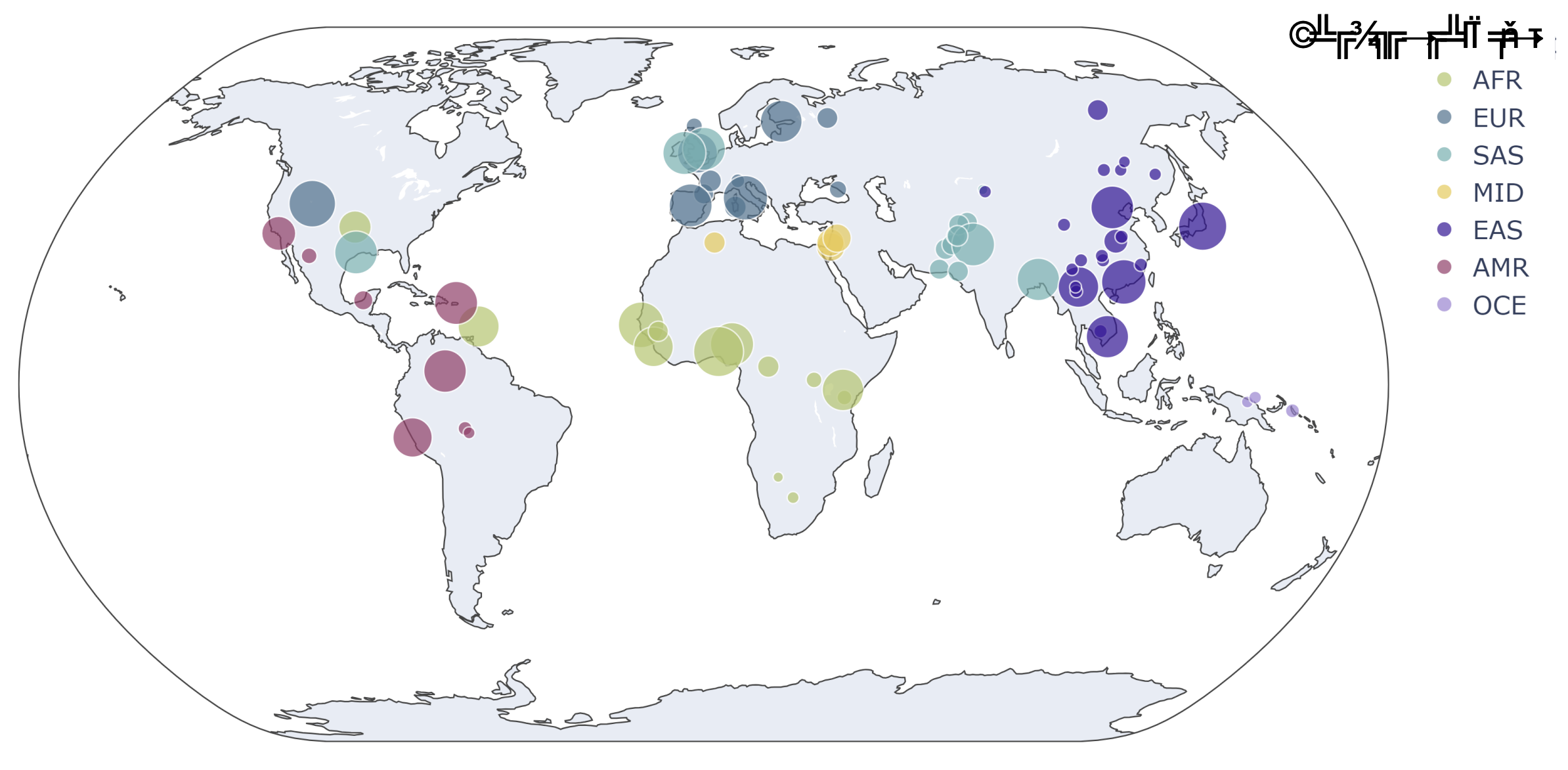
Comprehensive meta-analysis reveals the impact of human genetic variation on CRISPR off-target landscapes

Alyssa Ferreira, Senior Bioinformatics Engineer at MaxCyte
Computational methods alone are insufficient to fully assess the off-target risks of gene therapies. Biochemical and cellular assays are essential for comprehensive evaluation of gene editing specificity. Off-target assays are typically performed using only the reference human genome. Such single-sequence approaches entirely overlook human genetic diversity and may lead to the omission of variant-specific off-target sites. Genomic sequence variation may alter editing at the on-target locus or off-target loci identified in the reference genome or might give rise to entirely new off-target loci. This underscores the need for highly sensitive methods to thoroughly characterize off-target activity with a variant-aware population-scale approach. In this study, we present a meta-analysis that showcases the impact of human variation on the off-target activity of 84 guide RNAs (gRNAs). For 16 gRNAs, computational enumeration of putative off-target loci is complemented by biochemical data collected from the ONE-seq (Oligonucleotide Enrichment and Sequencing) assay. The ONE-seq assay leverages in vitro cleavage reactions using synthetic oligonucleotides, enabling collection of biochemical cleavage data of variant-specific, putative off-target sites across any known human genetic variation. This approach decouples assessment of biochemical cleavage from the availability of relevant genetic material and can facilitate scalable assessment of off-target risk across the intention to treat or global human populations. We observed that the inclusion of genetic variation increases the putative off-target space by an average of nearly 50%, corresponding to an average of almost 73,000 additional putative off-targets per gRNA. ONE-seq analysis demonstrated that over 30% of cleaved off-target loci were from variant genomes. We also found that 40% of the 84 gRNAs had variation at their on-target loci, with potential consequences for critical sequence elements such as the PAM and seed regions, which may affect editing efficiency. Furthermore, we explore the added value of integrating various publicly available resources for more complete representation of the human genome, including use of a pan-genome and a telomere-to-telomere (T2T) genome. The results presented here clearly underscore the necessity of considering human genetic variation during off-target assessment of the editing system – gRNA and nuclease. We propose a strategy that integrates variant-aware computational screening with scalable biochemical assays like ONE-seq to improve the design and safety evaluation of therapeutic gene-editing systems.