Scientific Brief
Base Editing of the Fetal Hemoglobin Promoter Yields Elevated Levels of HbF Expression in HSPC-Derived Red Blood Cells
Background
Hemoglobin is a multi-subunit protein responsible for oxygen transport in red blood cells. Hemoglobin consists of two α-globin subunits and either two γ-globin subunits (in fetal hemoglobin) or two β-globin subunits (in adult hemoglobin). After birth, the expression of γ-globin is gradually replaced by β-globin expression, until only adult hemoglobin is present.
Sickle cell disease is a group of blood disorders caused by different single-point mutations in the HBB gene encoding the β-globin subunit of hemoglobin. Mutant β globin polymerizes, causing red blood cells to acquire their hallmark “sickle” shape. Sickle cells hinder blood flow and have reduced longevity, causing symptoms ranging from tiredness and anemia to severe acute and chronic pain, immunodeficiency and multiple organ failure or stroke.
Various therapeutic approaches have been developed to treat sickle cell disease, focusing most recently on correcting the pathogenic point mutation in the patient’s HBB gene or by modifying the γ-globin (HBG) promoter to restore γ-globin expression and fetal hemoglobin (HbF) production.
Recently, Ravi et al. used an innovative base editing approach to screen the HBG promoter, identifying new point mutations that can induce γ-globin expression.1 This case study demonstrates the therapeutic potential of mutating a TT cluster at -123/124 in the HBG promoter in CD34+ HSPCs by targeted base editing. MaxCyte® electroporation enabled the efficient delivery of gRNA and adenine base editor mRNA to primary human HSPCs. Editing restored HbF and repressed HBB gene expression without impacting erythroid differentiation or enucleation.
Workflow

Results
Base editing at the novel cluster in the HBG promoter of HSPCs induces γ-globin expression and represses β-globin expression, restoring HbF in red blood cells.
CD34+ HSPCs were electroporated with adenine base editor mRNA (ABE8e) and gRNA targeting the known BCL11A site at position -115 (gRNA 2) or the novel cluster at position -123/124 (gRNA 11) in the HBG promoter, or with a control gRNA targeting the AAVS1 locus. After resting, cells were expanded and then cultured for erythroid differentiation and analyzed as indicated in the workflow.
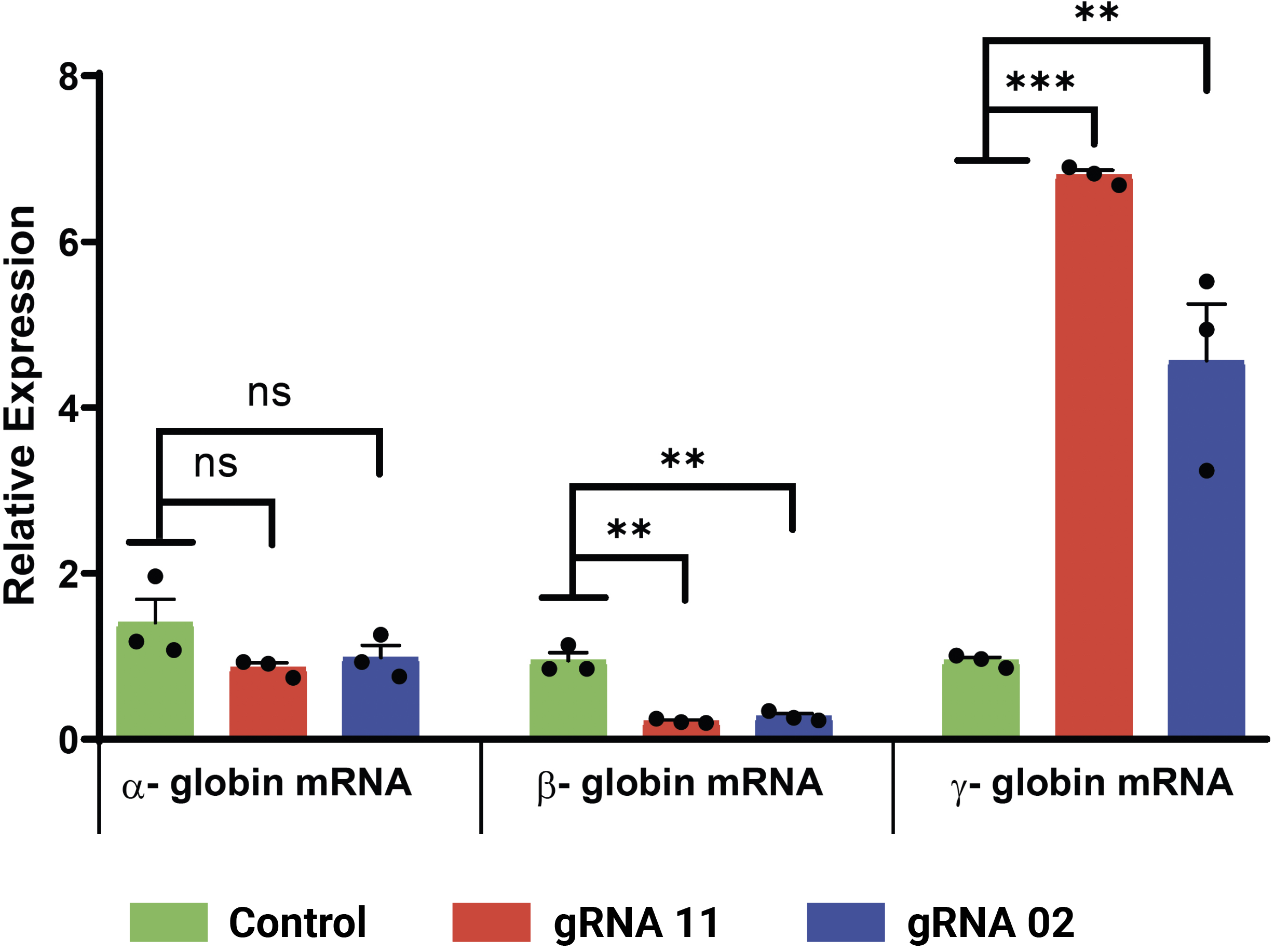
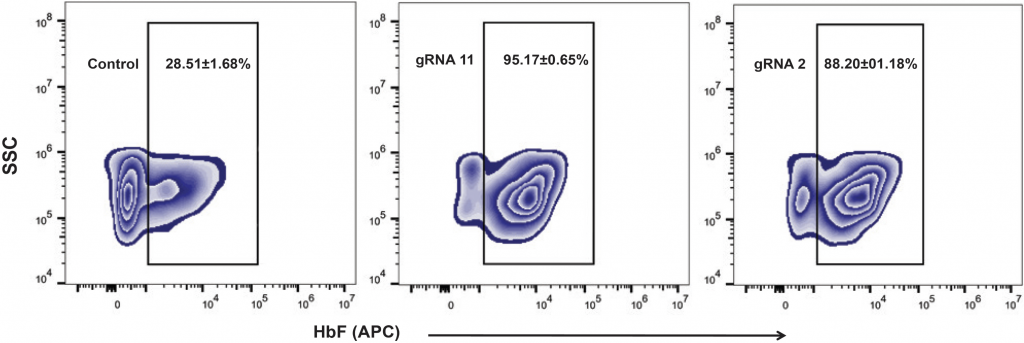
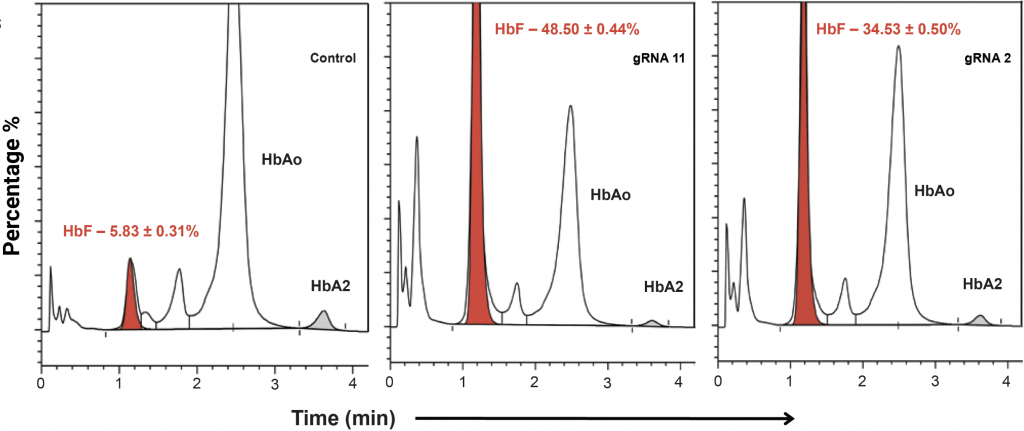
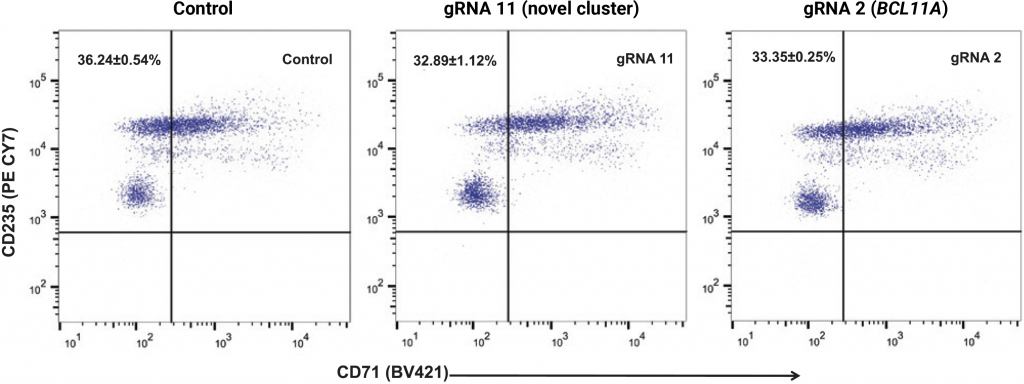
(A) Relative levels of α-, β- and γ-globin transcripts in differentiated CD34+ HSPCs (day 9) were assessed by qRT-PCR. Relative to the control, editing with gRNA 11 (> 6-fold induction) or gRNA 2 (> 5-fold induction) induced HBG transcription and repressed HBB transcription without impacting levels of α-globin transcripts. HbF flow cytometry (B) demonstrated that >95% of cells edited with gRNA 11 were HbF-positive. HPLC analysis (C) showed that nearly half of the hemoglobin in cells edited with gRNA 11 is HbF. Importantly, the enucleation (D) and expression of erythroid maturation markers (E) was not significantly impacted by base editing of the HBG promoter.
A) Relative levels of globin subunit transcripts
B) Proportion of HbF-positive cells
C) Ratio of Hb variants in differentiated cells
D) Enucleation pattern in differentiated cells
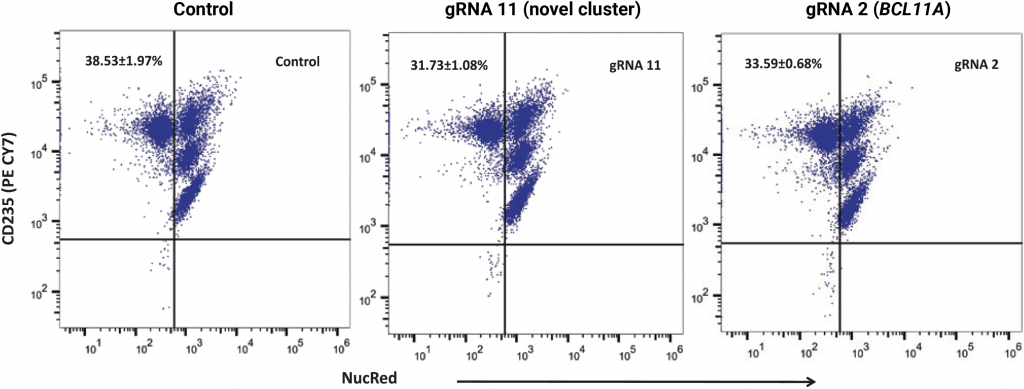
E) Expression of erythroid maturation markers (CD235a and CD71)
Summary
- MaxCyte electroporation efficiently delivered gRNA and base editor mRNA to CD34+ HSPCs.
- Adenine base editing at the -123/124 positions in the HBG promoter induced γ-globin transcription, restoring fetal hemoglobin expression.
- HBG promoter editing at the -123/124 cluster resulted in higher levels of fetal hemoglobin expression than editing at the known BCL11A (-115) site, demonstrating the therapeutic potential of this novel site.
Reference
- Ravi NS, Wienert B, Wyman SK, et al. Identification of novel HPFH-like mutations by CRISPR base editing that elevate the expression of fetal hemoglobin. eLife 2022, February 11. https://doi.org/10.7554/eLife.65421.
This content was adapted and reproduced from Ravi et al. eLife 2022;11:e65421. https://doi.org/10.7554/eLife.65421 under a Creative Commons Attribution License (CC BY 4.0).