Application Note
MaxCyte® Enabled Mammalian Display Method for the Development of Enhanced Affinity TCR-Based Biologics
Background
Cancer remains one of the leading causes of death globally, driving ongoing efforts to develop more effective treatments. T cell receptors (TCRs) are natively expressed on T cell membranes, where they recognize intracellular antigens that have been processed and displayed on target cells by human leukocyte antigen-1 (HLA-1). Because of their ability to recognize processed intracellular antigens, TCR-based cancer therapeutics (Figure 1) are emerging as an exciting alternative to bispecific antibodies or traditional CAR-T cells, which are restricted to antigens expressed on the tumor cell surface.
Principles of T cell receptor-based immunotherapy

Figure 1: A) T cells engineered to express a recombinant T cell receptor (TCR-T cells) engage with a specific tumor antigen displayed as a peptide:HLA-1 complex on tumor cell membranes. Once bound to their target antigen, TCR-T cells can produce cytolytic granules for tumor cell killing. B) Soluble bispecific T cell engaging receptors (TCERs) with a T cell receptor domain and a T cell binding domain bind their specific HLA-displayed target tumor antigen and recruit circulating T cells for a potent anti-tumor response without T cell engineering.
Soluble, bispecific TCR-based T cell engagers are therapeutic proteins comprising a TCR like domain that binds tumor peptide antigens displayed by HLA-1 (pHLA) and a domain to bind and recruit circulating T cells, typically based on an anti-CD3 antibody. If the naturally low antigen affinity of TCRs can be enhanced, T cell engagers have great potential as cancer immunotherapies.
Yeast or phage display can screen millions of sequences to identify TCR variants with high antigen affinity. Unfortunately, TCR affinities can drop when those higher affinity TCR sequences are expressed for clinical manufacturing in mammalian cell lines with mammalian post translational modifications. The mammalian display technique may be a better alternative, enabling the selection of high-affinity sequences in the final TCR-based bispecific context, avoiding reformatting or switching cell hosts for therapeutic manufacturing. Additionally, mammalian display may naturally select candidates with reduced immunogenicity, and favorable biophysical properties and developability.1
Mammalian display involves creating a library of mammalian cells that express variants of a candidate protein and display them on the cell membrane. The cells are then screened for antigen binding, and individual clones are selected, expanded and sequenced. There are various methods for mammalian display cell library creation, including gene editing techniques that can ensure the integration of one candidate sequence per cell. In this case, high-efficiency transfection of large numbers of cells is essential to create cell libraries with comprehensive representation of sequence variants. MaxCyte’s Flow Electroporation® technology can transfect up to 1x1011 cells in one cycle, reportedly enabling the generation of mammalian display libraries with up to 1x108 sequences.2
Aim
Researchers at the University of Stuttgart and Immatics Biotechnologies developed a mammalian display platform to select affinity-enhanced TCR sequences expressed in a proprietary bispecific T cell engaging receptor format, TCER®.3 The method was validated by testing the ability to identify enhanced affinity variants of an anti-PRAME (preferentially expressed antigen in melanoma) TCER. MaxCyte’s electroporation technology enabled the generation of a TCER CHO display library used for candidate selection and the transient expression of soluble, bispecific TCERs for functional characterization.
Methods
Workflow for mammalian display library generation and selection of affinity-enhanced TCERs

Landing Pad RFP_A03 CHO cells were electroporated with a PRAME TCER plasmid library and Flp recombinase mRNA. Following electroporation, TCER-expressing cells were expanded and sorted through positive selection with PRAME pHLA, followed by rounds of selection combined with counter-selection against PRAME-similar peptides to eliminate off-target binding. Individual clones were sequenced.
CHO display library generation
The researchers used recombinase-mediated cassette exchange (RMCE) in Landing Pad RFP_A03 CHO cells to create a mammalian display library of CHO cells expressing variants of a membrane-anchored anti-PRAME TCER (Figure 2). Landing Pad RFP_A03 CHO is a high-expression CHO cell line with a RMCE-targetable landing pad.3,4
Format of membrane-anchored TCER

Figure 2: One TCER chain comprises the variable TCR alpha chain (Vα), an anti-CD3 variable light chain domain (VL), IgG1 constant domains (CH2 and CH3) with knob-forming mutations and ablated Fc gamma receptor binding and complement activation, and a PDGFR-derived membrane anchor. The other TCER chain comprises an anti-CD3 variable heavy chain domain (VH), the variable TCR beta chain (Vβ), and IgG1 constant domains (CH2 and CH3) with hole-forming mutations and ablated Fc gamma receptor binding and complement activation.
Previously, 48 sequence variants in the complementarity-determining regions (CDRs) of a model anti-PRAME TCR were identified by yeast display. A plasmid library was constructed with over 36,000 TCER sequences representing all combinations of the previously identified CDR variants. The TCER plasmids contained a TCER expression cassette, encoding each TCER chain under the control of its own HCMV promoter, flanked by Flp recombinase target sites, FRT and FRT F3 (Figure 3a).
Landing Pad RFP_A03 CHO cells contain an RFP expression cassette flanked by Flp recombinase target sites, FRT and FRT F3 (Figure 3b). The presence of two different Flp recombinase target site sequences enables RMCE with a donor plasmid containing an identical arrangement of target site sequences (Figure 3c). Landing Pad RFP_A03 CHO cells were electroporated with an anti-PRAME TCER plasmid library and Flp recombinase mRNA to create an anti- PRAME mammalian display CHO cell library.
Creation of a mammalian display library by recombinase-mediated cassette exchange
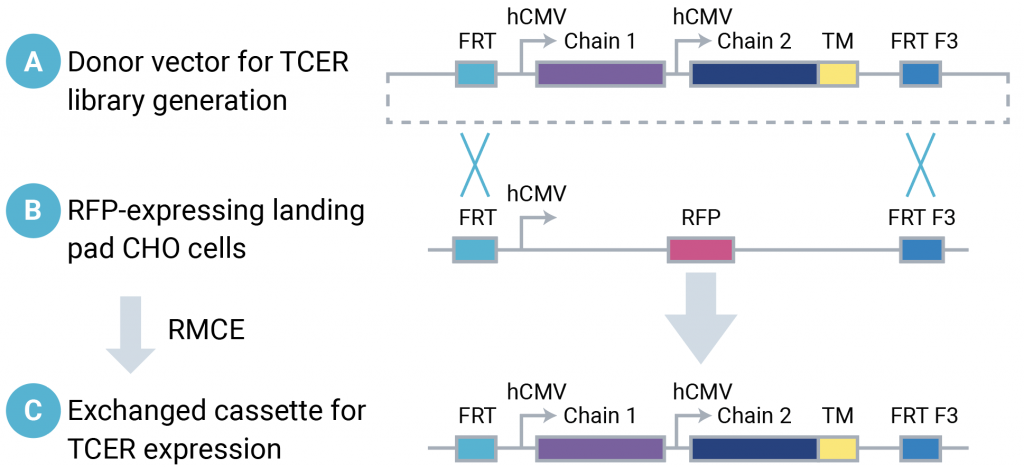
Figure 3: Donor plasmids containing a TCER expression cassette flanked by Flp recombinase target sequences (A) were introduced into Landing Pad RFP_A03 CHO cells containing the same arrangement of Flp recombinase target sequences (B), enabling recombinase-mediated cassette exchange to generate CHO cells containing the TCER expression cassette (C).
Selection of enhanced affinity PRAME TCER sequences
Following electroporation, TCER-expressing cells were sorted and expanded through a round of positive selection with PRAME pHLA (100 nM), followed by selection with PRAME pHLA (31.6 nM) with counter-selection against a pool of 11 PRAME-similar pHLA tetramers (10 nM each), then finally single-cell sorting with PRAME pHLA (10 nM) and 11 pooled PRAME-similar pHLAs (10 nM each). CHO cells expressing TCERs with high affinity for the target peptide are retained, while CHO cells expressing TCERs with off-target binding to similar sequences are excluded. Individual clones were sequenced.
Soluble TCER expression
Native TCRs are membrane-bound proteins with low stability when expressed as soluble molecules.5 This study aimed to develop a mammalian display platform to enable the discovery of therapeutic bispecific TCR-based TCERs. To assess the ability of this mammalian display method to identify functional, soluble TCERs, MaxCyte electroporation was used to produce the TCERs and the parental TCR by transient expression in CHO-S cells (Figure 4). Binding affinity and biological function of the soluble proteins was assessed.
MaxCyte workflow for soluble TCER expression

Figure 4: CHO-S cells were transfected by MaxCyte electroporation with TCER chains expressed from separate constructs (1:1 ratio). Cells were rested for 40 minutes. Media was added (4x106 cells/mL) and culture continued overnight (37°C). After 24 hours, the culture temperature was lowered to 32°C and sodium butyrate was added (final concentration 1 mM). Production cultures were fed on days four, six and eight. Protein was harvested on day 11, or when production culture viability dropped below 70%, and purified.
Results
Mammalian display identified enhanced affinity anti-PRAME TCER candidates
Analysis of 171 clones selected by mammalian display identified 39 unique TCER sequences. Ten of the most abundant sequences were chosen for further study. Flow cytometry assessed the on- and off-target binding of the 10 selected CHO-displayed TCER candidates. Six out of 10 candidates bound avidly to the target antigen, with a detectable signal at a concentration of 100 pM PRAME pHLA (Figure 5a).
The TCER candidates bound minimally to eight of the 11 PRAME-similar pHLA tetramers but bound with varying, stronger affinity to the three remaining sequences (Figure 5b). Although off-target binding was not eliminated, the authors believe this may reflect issues with the parental TCR sequence used in the previous yeast studies. Nonetheless, these results confirmed that this mammalian display method can identify TCER sequences with enhanced affinity target binding and demonstrated how the technique can be used to simultaneously counter-select for off-target binding.
On and off-target binding of TCER candidates

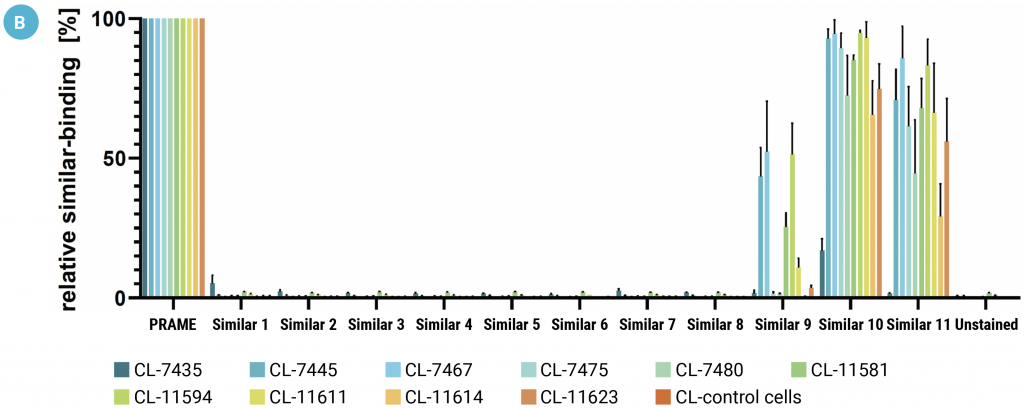
Figure 5: The binding characteristics of CHO-displayed TCERs were assessed by flow cytometry. A) Cells were incubated with PRAME pHLA monomer at 100 nM to 10 pM concentrations. B) Cells were incubated with 10 nM of a PRAME-similar pHLA tetramer as indicated. Each data point represents the mean of triplicate measurements with the respective SD.
Soluble TCER candidates retain enhanced affinity binding to PRAME pHLA
The binding affinity of soluble candidate TCERs to PRAME pHLA was determined by biolayer interferometry. The trend in binding affinity seen with CHO-displayed TCERs (Figure 5a) was broadly mirrored with the soluble TCERs. All the TCERs tested bound the target antigen more strongly (KD from 3.4-37.4 nM) than the parental TCR (KD 1230 nM) with the best candidate—CL-7467—showing about a 400-fold increase in binding affinity over the parental TCR.
Soluble TCERs are biologically active
The anti-tumor activity of the soluble TCERs was tested against target cells presenting different concentrations of PRAME pHLA and against PRAME pHLA negative SET-2 cells (Figure 6).
TCER functional characterization

Figure 6: Healthy donor PBMCs were incubated with either UACC-257, HS695T, A375 or SET-2 target cells (E:T ratio of 10:1) with increasing concentrations of soluble TCER candidates (0.01 pM to 10 nM). Supernatants were analyzed for cytotoxicity and cytokine release after a 48-hour co-culture.
TCER-dependent cytotoxicity is antigen-specific
LDH-release assays were performed against UACC-257 cells (approximately 1100 PRAME copies per cell), HS695T cells (400-500 PRAME copies per cell), A375 cells (approximately 50 PRAME copies per cell) and PRAME negative SET-2 cells. Cytotoxicity was broadly as expected based on PRAME pHLA display levels and TCER binding affinity (Figure 7). No killing or very weak cytotoxicity at the highest TCER concentrations was seen for all candidates against the SET-2 negative control (Figure 7).
TCER-mediated off-target cell lysis

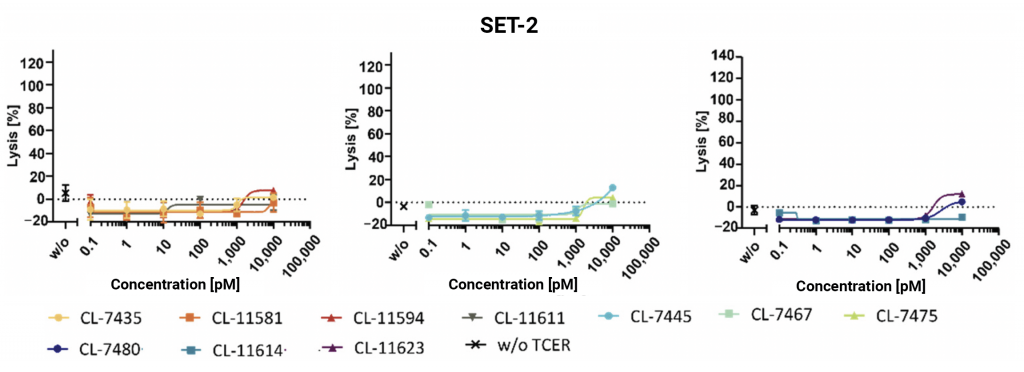
Figure 7: Healthy donor PBMCs were co-cultured with the target cell line indicated (E:T ratio of 10:1) and increasing TCER concentrations. Cytotoxicity was determined by LDH-release assay. Each data point represents the mean of triplicate measurements with the respective SD.
TCER-dependent cytotoxicity is antigen-specific
TCER-mediated cytokine release from healthy donor PBMCs incubated with three leads candidates and co-cultured with UACC-257 target cells was quantified (Figure 8). The three soluble TCER lead candidates all triggered IFNγ, IL-2, perforin and granzyme B release from healthy donor PBMCs following a tumor cell challenge. No TCER-mediated induction of IL-2 or perforin release was observed when PBMCs were co-cultured with the SET-2 negative control cell line.3
TCER-mediated cytokine release by PBMC in co-culture with tumor cells
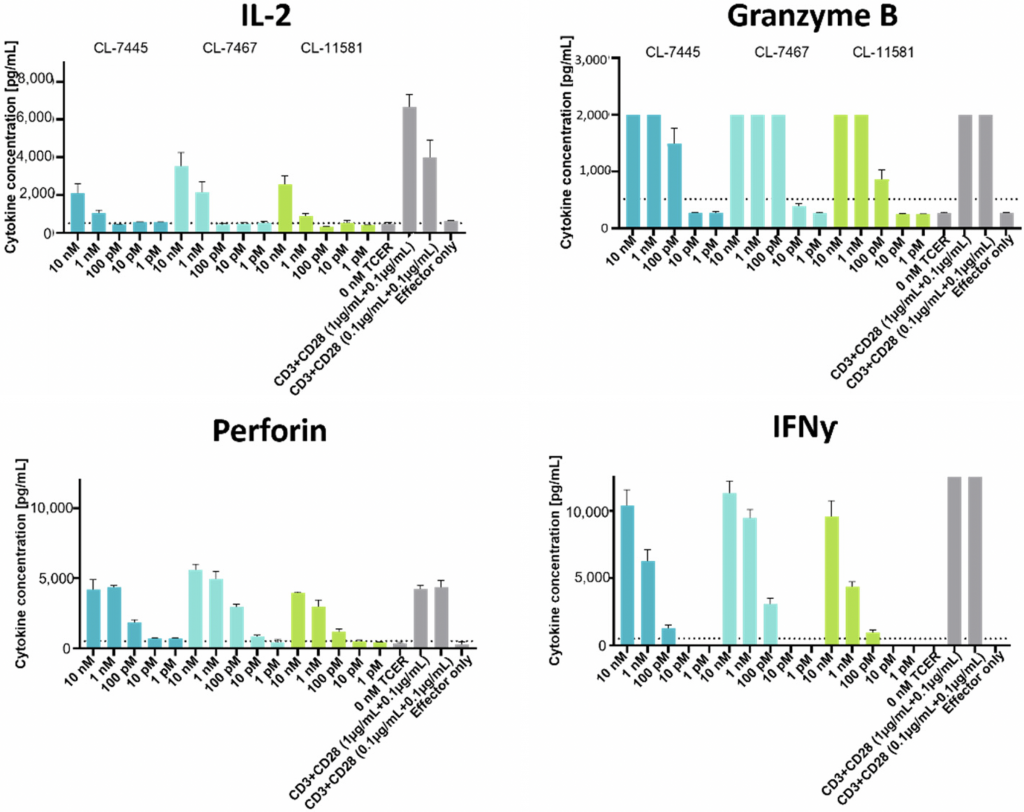
Figure 8: Healthy donor PBMCs were co-cultured with UACC-257 target cells (E:T ratio of 10:1) with increasing concentrations of the three leading TCER candidates. CD3- and CD28-binding antibodies were used as positive control. Cytokine levels were determined using a cytotoxic T/NK cell kit. Each data point represents the mean of triplicate measurements with the respective SD.
Conclusion
This study described the development and validation of a CHO-based mammalian display platform for identifying enhanced affinity bispecific T cell-engaging receptors, TCERs. Mammalian display has several advantages over yeast or phage display methods, including reduced immunogenicity and favorable developability when selecting candidate therapeutics. Lead discovery in the therapeutic bispecific TCER context avoids the need for reformatting, which could result in a loss of affinity or function.
Several gene editing techniques have been used to create mammalian display cell libraries, including transposase, nuclease and recombinase-mediated integration, with reported integration efficiency ranging from 0.5 to 7.5%. High integration rates are essential to ensure the display cell library accurately represents the starting sequence library. Hence, transfection efficiency and scale are critical factors for successful mammalian display screening.
The efficiency, viability and scale of MaxCyte electroporation can theoretically enable the generation of mammalian display libraries containing up to 1x109 sequences. MaxCyte electroporation is a versatile transfection technology that enabled the development of a Flp landing pad CHO cell line, the generation of a TCER CHO library, and the production of biologically active soluble TCER candidates by transient gene expression.
References
- Dyson MR, Masters E, Pazeraitis D, et al. Beyond affinity: selection of antibody variants with optimal biophysical properties and reduced immunogenicity from mammalian display libraries. MAbs. 2020;12(1):1829335. doi:10.1080/19420862.2020.1829335M
- Slavny P, Hegde M, Doerner A, et al. Advancements in mammalian display technology for therapeutic antibody development and beyond: current landscape, challenges, and future prospects. Front Immunol. 2024;15:1469329. Published 2024 Sep 24. doi:10.3389/fimmu.2024.1469329
- Dilchert J, Hofmann M, Unverdorben F, Kontermann R, Bunk S. Mammalian Display Platform for the Maturation of Bispecific TCR-Based Molecules. Antibodies (Basel). 2022;11(2):34. Published 2022 May 10. doi:10.3390/antib11020034
- MaxCyte Scientific Brief describing the development of Landing Pad RFP_03 (SB-24382)
- Robinson RA, McMurran C, McCully ML, Cole DK. Engineering soluble T-cell receptors for therapy. FEBS J. 2021;288(21):6159-6173. doi:10.1111/febs.15780






