How to Mitigate Gene Editing Program Risk through Comprehensive Off-Target Safety Profiling & Characterization
Papillon Therapeutics
Webinar
June 24, 2025
Papillon Therapeutics and SeQure DX, a MaxCyte company, discuss how to strengthen the safety of your CRISPR-based therapeutic program through in silico and in vitro assessments
Are you engineering a CRISPR-based editing system for application in cell or gene therapy? Learn how to assemble an off-target safety package for your IND submission. For those in earlier stages of development, learn how early assessment of off-target risks can help de-risk your program.
Key topics covered
-
Guide selection approaches for the discovery stage: Identifying off-target effects remains a critical challenge in gene editing. SeQure DX's two-step, variant-aware assay approach helps you find gene editing off-targets safely and efficiently, setting up your program for regulatory approval down the line. Learn about the latest tools used to generate off-target data for both CRISPR/Cas and base editing systems.
-
Gene therapies for HSPCs: Papillon Therapeutics presents their program on gene-modified and gene-edited hematopoietic stem and progenitor cells, or HSPCS.
-
The path to IND submission for Friedreich's ataxia study: Papillon Therapeutics shares their experience collaborating with SeQure DX to advance a Friedreich’s ataxia program toward IND submission, including a preview of their off-target safety data package.
Watch how to evaluate off-target effects and reduce gene editing risk

Presenters



Have more questions?
Send your question to one of our cell engineering experts.
Related resources
Transcript
Vijetha Vemulapalli: All right. Welcome everybody. Good morning or good afternoon, depending on whichever time zone you are in. Thank you for your interest in attending the webinar. We'd be happy to take you through some slides to talk about different ways to evaluate off-target effects. And we have a couple of guest speakers here that are doing some really good work with Friedreich's ataxia, and they're going to present some of the off-target work there as well.
This is just a disclaimer slide in terms of agenda. I can get us started off providing an overview about off-target evaluation and then a little bit about guide selection and products we have for the discovery stage. Then I'm going to hand it off to Stephanie. Stephanie Cherqui is a professor at the Department of Pediatrics at University of California San Diego. She's also the director of the Gene Therapy Initiative and co-founder of Papillon Therapeutics. We also have Colin Exline, he's going to talk about the path to IND for the Friedreich's ataxia program, and he's the director of research and development at Papillon Therapeutics.
So diving right in. As many of you know, there are multiple gene editing platforms that have been described and are being deployed as therapeutics. So on the left side of the slide, you can see the original targeted nucleus editors. These are really well-known, and most well-known of these are RNA-guided CRISPR-Cas nucleuses.
More recently, there have been, what we're calling CRISPR 2.0 editors. These are based on the original CRISPR-Cas nucleuses, but they're a little bit more precise where base editors can induce specific nucleotide substitutions in a targeted way, while primes can essentially make any nucleotide substitution, or even small insertions or deletions at the desired locus in living cells.
So we essentially have the capability of making any small, genetic change we want. But one of the important challenges in the field, as you are thinking about advancing the use of these gene editing modalities into therapeutics is understanding where else in the genome these editors may be causing unwanted, changes, which we are calling off-targets.
And because these are editors, we're making permanent off-target mutations, right? So there've been several challenges for the field. One of the biggest things is that there's no gold standard that exists for off-target determination. Partly this is because whole genome sequencing is not cost-effective or sufficiently sensitive.
But it’s also because it's a very rapidly moving field. There've been a variety of off-target assays that have been developed to date, but every one of them is experimentally challenging to do. They also require specialized deep computational expertise in order to be able to analyze and interpret the results and basically look at the data that comes out of these assays.
So overall the field has settled on a consensus of doing a two-step approach, to find these off-targets. The first step is what we're calling nomination, and the second step is what we're calling confirmation, or sometimes you might hear it being called verification.
The first step, nomination, what we are trying to do is identify potential off-target sites that occur in a genome. The sensitivity in this step is really, really important because it defines which sites will be examined in the subsequent confirmation step. So in this figure here, if you're looking at the left side, if you think of the genome as this dark space and the Xs are representing off-target sites that occur within the genome, the nomination assay is used to identify potential off-targets, which is basically shedding light on certain parts of the genome. What you want to make sure is if those Xs are the off-targets, you're shedding light on every single one of those Xs. And if you do shed light on other regions, it's okay but you really don't want to miss any of the Xs.
There've been many different types of nomination assays that have been described to date, and we'll talk about some examples here, but broadly speaking, they're either in silico or in cellulo or cell-based assays or in vitro or biochemical assays. But because there's really no gold standard, we should ideally use orthogonal methods to nominate as many off targets as possible and identify a “super-set” of off-targets and move them into confirmatory steps.
With the confirmation, which is on the right side of the slide, what we want to do is look at these nominated sites in an actual editor-treated target cell or tissue, or maybe an appropriate surrogate set of cells—and not all the nominated sites will be confirmed as being bona fide off-targets in the therapeutic or cellular context. This can be because of many reasons, right? It could be the epigenetic status of the target sites, maybe it's the expression level, the dose, the duration or how the gene editing is done. Also when you're looking at confirmation, we should not only look for substitutions, but also at small insertions and deletions and structural variations as well. And we'll address this as we move forward.
So first I want to walk you through a couple of different nomination assays. The first type of assay you can use is an in silico approach, and broadly speaking, there are a couple of different categories. One would be where you're generating a list of closely matched sites in the genome. We know there are several tools that can do this, and there's some examples listed on the slide. This is because we know that the off-target sites don't occur randomly. They occur at sites that resemble the on-target site. The problem with this approach, though, is that there are so many different sites that you can identify and even if you go to a small number of mismatches, which the table on the right shows, you can end up with a really large number of potential off-target sites. And this is just too many for the actual confirmation assays. The second way to do in silico methods is to use algorithms that help you identify which closely match sites are likely to be bona fide off-targets.
But the problem is these programs miss known bona fide sites. So the conclusion from all of this is that the in silico assays alone are really not sufficient to be an effective nomination tool. You do need to, at the very least, supplement or, better yet, replace them with experimental nomination methods instead of the in silico methods.
Another class of nomination assays are cell-based assays. The most widely used cell-based assay to date is the GUIDE-Seq assay. This came out of Keith Joung’s lab. At this point we believe it's essentially anyone that's moved into IND has used it as part of their off-target safety analysis. The way that the GUIDE-Seq assay works is you deliver the editor you're interested [in] into cells, so you introduce a short double-stranded oligo nucleotide tag that basically has a sequence that's orthogonal to the human genome. You can prime against that sequence without any link to your primer to other sites of the human genome. What happens then is that the editor creates double-stranded breaks, and this double-stranded oligo jumps in and gets ligated. It essentially tags all the sites in the genome in which you have the break occurring. So you can take the genomic DNA from these edited sites and prime against the oligo tag and do one-sided amplification and then do next-generation sequencing as a way to nominate all potential off-target sites in the genome. It can work for pretty much all gene editing nucleases, although it depends on the type of cut that's introduced.
These cell-based assays have both advantages and disadvantages. One advantage is that you can nominate sites in a cell type that's relevant to the context of the therapeutic program, but the flip side is that you may not be able to nominate in the most relevant cell types. For example, it's known that GUIDE-Seq shows toxicity in IPSCs, so it limits the type of cells that you can do the assay in. There are some potential sensitivity limitations as well because you're working in cells. Things like fitness can be an issue, especially if you are expressing the nucleus, because you're trying to drive sensitivity and find all the potential off-targets in these cells.
So moving forward and looking into another class of nomination assays, which are the in vitro biochemical ones. What we mean is that the assays are performed basically in a test tube with purified components. We believe that ONE-Seq is one of the most highly sensitive in vitro assays that has been described to date. The way that ONE-Seq works is that, initially, you do the same type of in silico computational enumeration to list out all the closely matched off-target sites in the genome. You can do this on one genome, but you can actually do it in as many genomes as you want, and then you use, high-density oligonucleotide synthesis to create a library that represents all of the generated off-target sites. Each site is encoded with a unique barcode, so it can be tracked and counted. And then you subject this library to ONE-Seq selection. And again, you can do this with all the nucleuses, you can do it with different base editors. It can be adapted to prime editors as well. And then essentially, the potential off-target sites are cleaved and you can ligate sequencing adapters to the cleaved sites, sequence the barcode and determine which of the sites were actually cleaved. We think that ONE-Seq is a great example of an in vitro assay because it's highly sensitive compared to the other assays that have been described, whether it's nuclease base or prime editor.
Now one of the big advantages of ONE-Seq is that you can take into account human genetic variation. Why is this important? Editors are essentially sequence-specific recognition agents, so they recognize a specific target site but they're also going to recognize off-target sites that are closely related. But then none of our genomes are the same. We can have small nucleotide changes and other differences in the genome that can really influence what the off-target profile looks like from individual to individual. So ideally, you want to be able to do these nomination assays on a wider representation of individuals, so you can account for more common genetic variations as you nominate off-targets.
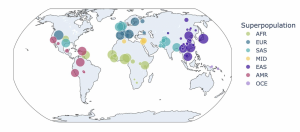
The nice thing about ONE-Seq is that you can do the initial computational enrichment on more than one genome. With the SeQure assays, by default, we look at the 1000 Genomes and HGDP [Human Genome Diversity Project] data sets, which encompass about 3,500 individual haplotype phase genomes that represent all seven major super populations. In a singular reaction, you can essentially assess all of the potential sites, whether they're edited or not, for a given editing system. We’re calling this variant-aware nomination.
In contrast with every other off-target nomination assay, it's really easy to scale across multiple genomes because you only need to do the assay one time. For example, if you were looking at 3,500 different genomes and you were using a different assay, you would have to repeat the assay multiple times to make sure all the genetic variation is encompassed in the assay.
So that was a little bit about nomination assays. Moving forward into step two, which is looking at the confirmation assays. You ideally want to perform the assays on therapeutically relevant cells, or at least a surrogate cell line that's therapeutically relevant. As I mentioned before, there are two types of edits you want to look at. The classic edits are just small indels. You can use targeted Amplicon sequencing and then next-generation sequencing to do this.
One of the challenges is that the amount of genetic DNA that's available to use as a template, particularly if you're looking for hundreds and thousands of these potential target sites, can be limiting. So using multiplex methods like Amplicon sequencing, rhAmpSeq or hybrid capture, these other methods are more practical.
The other things you want to make sure you do is also to look at larger-scale genomic alterations because we know that both the off-target and the on-target sites can participate in deletions, inversions and translocations.
Alright, so this leads us to a high-level view of SeQure’s current portfolio. We have a couple of different offerings that are on the nomination side of things. There's the ONE-Seq assay, the DEUX-Seq assay, which is an adaptation of the Digenome-seq method. We talked about GUIDE-Seq earlier. We also talked about using applicant sequencing as a confirmation assay, and we also have a new method called SAFER Detection to look at large-scale genomic rearrangements. This is on the nomination and confirmation side. The presentations that Stephanie and Colin will be providing, they'll share some of the data that was generated from the nomination and confirmation methods.
I'm going to pivot and talk about screening. Let's say you are in an earlier phase of the program and you're in the process of identifying the right guide RNAs to move forward with. There are a couple of different products that can help you with that. Those are the Guide Profiler and Guide Select products. And I can go in depth into each of those.
First, I want to talk about the Guide Profiler product. This is entirely in silico work very similar to what is done for ONE-Seq. Going through the workflow, we perform a comprehensive computational search across multiple genomes, including variant genomes. One thing we do is to look for variants at the on-target locus. And then perform benchmarking for how the current guides perform against published or well-known guides. And then perform some annotation, which is basically assessing the functional impact for the potential off-target sites. And all of them are summarized and presented in a comprehensive report in both a PDF and an HTML format.
I'm going to move on to talk about some example figures and a sample data set that was generated using the Guide Profiler product. Here, we were looking to design guides targeting the TRBC gene. The off-targets were generated based on the hg38 genome as well as the 3,500 haplotype phase genomes I mentioned earlier.
The table on the left shows you a list of guides that were considered, the map in the middle shows you the representation of individual genomes across the globe that were considered for off-target assessment and then the bar chart on the right shows you for each of the guides that were considered—I think in this example there were 25 different guides that were considered—how many off-target sites can be identified, if you're doing a really comprehensive search, for up to six differences, allowing for indels, gaps between Proto and PAM. What is the off-target space looking like? The blue portion of the bar shows you the contribution of off-targets from the reference genome and the green portion shows you the contribution of off-targets from the variants. You can see that quite a few off-targets can arise from human genetic diversity, and this is the reason it's really important to consider that upfront.
Once all these off-targets are nominated, they're annotated with quite a few functional features. This includes proximity to the target sequence because closeness to the target sequence and high-edit frequency can increase the chances of rearrangements. We also look at the presence in cancer genes, proximity to disease genes, whether they're in genic or non-genic regions and any gene expression impact there might be. There's also annotation that looks at the effect of variation. Should that off-target site be edited? So all of this is really comprehensive. It's quite a bit of data to enable interpreting this data in an easy way. We condense all of that into a single numeric scale. We're calling this the “annotation concern score.”
The data that's presented in the report really provides quite a few figures and tables that help in interpreting the data. We’ve noticed that 40% of the guides that we've looked at to date have variation at the on-target locus, so it's really important to look at that early on during the guide selection. There are a couple of examples from the TRBC data set here. If you look at TRBC-20 ,there are a couple of different variants that are present, and if you look at the allele frequency table on the right, they're present at fairly low allele frequencies. However, the TRBC-25 guide has only one variant but it's present at a fairly high global allele frequency. So maybe that's something you want to take a closer look at to see if the variant can change the editing efficiency at the on-target locus.
Another example figure that could be in the report is proximity to the off-target loci, proximity of the off-target to the on-target loci. There are three different TRBC guides that are shown here, and you can really see how some of the guides are much cleaner compared to the others. TRBC-10, which is in the middle has basically no generated off-targets that are one to three differences away on the same chromosome, so the probability of large-scale genomic changes can be fairly low. However, with TRBC-1 and -11, you see off-targets that are close to the on-target locus. So there's increased chance of rearrangements. The table at the bottom just shows you how many off-targets are linked to cancer-causing and disease databases. So this is a fairly high-level summary. There are some additional tables that are present in the report as well.
But if you don't want to take a deep dive, there's a table that's presented at the very top of the report where all the functional features that are evaluated are shown. All of this culminates into a score, and then ranks are provided based on the score. So you can really start with a rank list of guides and then dive into the additional data if there is interest.
The advantages of using this tool are that you can really screen tens of guides at the same time to look at all of them side by side and identify a subset of guides to move forward with, and it's also in silico, so it's extremely fast.
Moving forward and talking about the Guide Select assay. Assuming you have a set of guide RNAs, let's say between two and eight guide RNAs, it makes sense to go beyond the in silico approach and collect some biochemical cleavage data to make some decisions on which guide to move forward with. The Guide Select assay is an efficient way to do this. It's basically a multiplex version of the ONE-Seq assay, so it's set up to collect in vitro cleavage data on a small subset of highly sequenced similar, putative off-target sites.
We went over, high-level, how the ONE-Seq works at the beginning of the presentation, but as a quick refresher, you start with the computational search, enumerate all the sequence similar off-target sites and then the oligos are synthesized to create a uniform ONE-Seq library. And then we perform the IVC reaction and then analyze to identify off targets that are actually cleaved. This is little bit of a side-by-side comparison of the Guide Select assay versus the full ONE-Seq. One thing to note is that we do keep the variant-aware capability of the ONE-Seq assay. You can look at up to eight guides at the same time, and we still do three replicates, the biological functional annotation is done, a really comprehensive report is done—and it takes about the same time.
The trade-offs that are done for multiplexing are to do a more limited in silico search where you look for only up to four differences, but with the ONE-Seq assay, you would look for up to six differences. So the number of sites that are expected per guide would be smaller, which is only up to 5,000 for Guide Select, but up to 244,000 for ONE-Seq. Also the IVC conditions, which is basically how much DNA do you have versus the nuclease and the guide, we perform it at different concentrations to collect more comprehensive data for the ONE-Seq assay. But for Guide Select, we would only do one dilution for efficiency's sake.
So what the Guide Select assay produces is very similar to the ONE-Seq assay. Here we're showing the off-target sites identified from the Guide Select assay along a couple of different dimensions. The ONE-Seq score on the y-axis and the annotation concern score on the x-axis, and each of those dots represent an off-target site that's cleaved at an appreciable frequency, so 0.001 of the on-target frequency. You want to pay attention to off-target sites that score high on one of the two dimensions: that would be Tier 1, 2a and 2b.
It shows you that just a quick look at this figure, you can tell that tier TRBC-22, which is the one on the left, has a much cleaner profile in relation to TRBC-15, which is on the right side of the slide. So all of this results in a really comprehensive report that's similar to the ONE-Seq report but showing a little bit more of a detailed view of the TRBC data that was generated.
In this example, the Guide Select assay was run on seven different TRBC guides and the summary table on the left shows you the number of loci that are screened by ONE-Seq and some of the high-level findings, such as how many were on-target proximal, how many of the nominated sites were present in variants with high allele frequency, and if they were in cancer-causing genes.
The guides are also colored based on how well they performed. You can see on the bar plot on the right side as well that the TRBC-22 guide had the cleanest profile followed by TRBC-10 and TRBC-5, and the rest of the TRBC guides had a much noisier profile. So if you were to use the Guide Select data, just based off of this view, it looks like there are three candidates that you can move forward with, with TRBC-22 being the most favorable one. Some additional data that's included in the report: we saw similar figures that show the proximity of high-frequency off targets that are close to the on-target locus. Again, 22 and 10 have really favorable profiles. However, with TRBC-18, there are three Tier 1 off-targets that are in proximity to the on-target locus. So, TRBC-18 is clearly risky in terms of structural rearrangements. Another example figure to the right shows you where the nominated off-targets come from, if they arise from the reference genome or from the variant genomes across the three different tiers used for nomination. Those are sunburst plots where the green parts show you what fraction come from variants and the blue parts show you all the sites that were nominated in a tier, regardless of their origin from the reference genome or the variant genomes. You can see clearly that for TRBC-22, based on the bar plot below, none of the variants are actually high allele frequency.
They're between zero and 0.01, so they're fairly low risk. However, in contrast, you see with TRBC-15, the bar chart shows there are several alleles, several variants with high allele frequency that is over 0.1. So these are some of the different pieces of information that are available to make some decisions on what the safest guide to move forward might be.
That brings me to the end of a methods and assays overview. I'm now going hand it off to Stephanie, who's going to talk about the application of gene editing to Friedreich's ataxia.
Stephanie Cherqui: Thank you very much, Vijetha. I will present our program, gene-editing hematopoietic stem cells for Friedreich’s ataxia, and Collin will then present our data with the ONE-Seq assays. I am a co-founder of Papillon Therapeutics.
What is the autologous hematopoietic stem cell transplantation? What do we do? We harvest hematopoietic stem cells from the blood after mobilization, and they are ex vivo and can be gene modified by adding a gene, using a lentivirus vector or other vector, or gene editing. Then they are tested and if they meet the release criteria, the patient goes through a chemotherapy to remove their own stem cells, and the cells are reinfused. This technology has a lot of advantages.
This approach has not been applied to many immune blood disorders, like immunodeficiency disease, but also the monocyte can integrate the tissues and they can give rise to macrophages or microglia in the tissues, and they have been used to treat tissue disorders, and also neurological disorders.
So in our group, we have used this ex vivo gene therapy for hematopoietic stem cells for cystinoosis, which is a multi-systemic, metabolic disorder. And we started this project in 2007 with a proof of concept. We have performed all the preclinical studies and then we have interacted with the FDA in 2012 and pre-IND. Then then we went through all the safety studies, manufacturing development. And as you can appreciate, this took several years before we got IND-cleared in 2018 to start a phase ½ clinical trial. We started this clinical trial in 2019, and we have enrolled over six targeted patients. Now this phase 1/2 is completed, and we are in the phase of a long-term follow-up of the patient. This program was sub-licensed by ABROBIO in 2017 and acquired by Novartis in 2023. And they just recently opened a phase for a pediatric population. As you can appreciate, this takes a long time. And in this case of cystinosis, we have added a gene with the lentivirus vector.
The program that I will talk now is about Friedreich's ataxia, a neurodegenerative disorder that starts in children, which affects locomotive ability. We have done a proof of concept, and we published our data in 2007. We showed that hematopoietic stem cells could completely rescue the locomotive ability of mice. And we have shown that the hematopoietic stem cells were able to engraft into the brain, dorsal root ganglia, spinal cord. They're also in the heart and muscle, but are also affected by the disease. And they're able to prevent neurodegeneration or cardiac dysfunction. And we have shown that the mechanism of action involves the transfer of the vaccine from the macrophage or microglia-derived cells to the neurons and myocytes.
In the case of Friedreich's ataxia, overexpression of frataxin is toxic, and most of the patients carry the same mutation, which consists of a GAA repeat expansion mutation in intron 1. So in this case, gene editing was much more applicable than adding a gene. At the beginning of this project ,we were lucky enough to collaborate with a group that in part chose a guide to cut this GAA mutation expansion in intron 1, so we'll call them UP4 and DN4. As Vijetha described, I want to stress the fact that we are very lucky that we chose guide RNA that were very efficient but very safe. As you can appreciate this takes many years to safely study, so we don't want, down the line, to realize that this guide was not safe enough to move forward to clinical trial. So we use this guide along with a Cas-RNA complex, and we electroporate hematopoietic stem cells to gene modify them. This approach has been approved for sickle cell disease and beta thalassemia with Casgevy by Vertex Pharmaceuticals.
Of course you have to do several steps for IND, and one part is the pharmacology studies. And for that we used the mouse model of Friedreich’s ataxia that carry the human FXN transgene with over 800 GAA trinucleotide repeats. We have shown that we were able to gene-modify and cut this expansion into hematopoietic stem cells from the murine model, and that all the hematopoietic stem cells that were gene corrected could be found in the hematopoietic tissues—but also in all the tissues affected by the disease. We also have shown that through this approach we're able to rescue the gait and balance in the mice and preserve the neurons at the granular cell layer. We also showed the improvement of cardiac function in these mice and the mitochondrial architecture and phenotype.
Moving forward, we then scale up our manufacturing process. For that, we did the process development, and we went ahead with two large-scale [runs] with this electroporation system. And we have shown in two different donors that we had good viability after electroporation of our RNP complex, and we were also reaching about 20% gene editing efficiency, which we have shown in the mouse model that this was at therapeutic level.
When we did our INTERACT meeting with the FDA, the FDA tentatively agreed with our setup and proposed plan experiment for the IND, but they really stressed the fact that they wanted a comprehensive off-target assessment to show the safety of our guides and to show also that there was no chromosomal rearrangement.
This was the time we reached out to SeQure because of their previous experience with IND and with other groups. We showed the data that we obtained with ONE-Seq. They also proposed, as Vijetha said, SAFER, which is to determine the large genomic rearrangement. We have had a really good experience with SeQure and we are also complementing our off-target and chromosomal rearrangement study with KromaTiD and IDT. So now, Colin will move on to show you our data with ONE-Seq.
Colin Exline: Thanks, Stephanie. As you said, we reached out to SeQure very early on to analyze our off-target profile and really dig into our safety studies to verify that this therapy should be moving through the IND pathway. We reached out and asked them to do the in silico analysis that was described earlier, utilizing six mismatches with the different characteristics shown here on the left for each of our two guide RNAs.
Remember, these have to be delivered in concert. And when we speak about gene editing, what we're speaking about in this instance, too, for our purposes, is the excision of the whole GAA repeat region. So while we are getting 20% in those areas, each of these gRNAs themselves are getting 70, 80, 90% in our target populations. So it was really important to know I have a very thorough analysis of what could be a putative, non-targeted cut in our system.
And this is what we got back. We were really happy because our initial analysis looked really clean and then we get 50,000 possible loci that are off-target sites. And you can see the breakdowns here. Fortunately, most of them were more than, say, one mismatch and even up to none under three in the UP4. We took those, as has been said, synthesized them in vitro and then ran the ONE-Seq assay. I wanted to give you a brief example of the kind of data that you get back from this.
Here's the table from one of our guide RNAs. What you can see is it gives you genomic position, it gives you gene names, go terms, links out to public databases for effects. Where that is located in a gene, so is this in a coding sequence? Is it intronic? Is it in an upstream region, a downstream region, untranslated region, what have you. What the predicted outcome of an editing event at that would be, so if it's an intron, is it just some sort of a new intronic variance or are you going to possibly have frame shifts within your coding sequence?
And going through, looking at what cleaved in an in vitro assay, so like this idealized situation greater than 0.01%, we were able to narrow down our list of 50,000 genes to a handful, less than 200 at each site, which was really beneficial to helping ease some of our concerns as we were moving forward. We felt really good at the beginning, and then we got this list of 50,000 genes and got a little more concerned. Now we're looking at a lower level. I think this also helps put this into perspective because you can see what other people are obtaining and where you kind of slot in for possible numbers of off-target sites, and we were very happy to see that we're right within the ranges of some very well-described sgRNAs.
On the right here, I think it also is another testament to why you need to look at the multiple variants because if you look at our UP4, the tier 1 concern score category, we would've missed that entirely possibly if we didn't have that variant. And so it's something we need to be aware of going forward.
We did take this with us to our pre-IND meeting, and they had very favorable viewpoints towards this variant-aware profiling of our off-target as one of our orthogonal methods for demonstrating off-target safety.
As Vijetha said earlier, sometimes you get surprised and your on-target has a variant sequence within it. And that actually did occur here. So we have two putative variant SNPs within our on-target site for one gRNA. you can see here the frequency with which these are found in the population. So on all individuals, less than 0.2%, and even in specific populations, less than 1%. So not something that we necessarily have to be horribly concerned about. Additionally, when they, and I didn't point this out on the ONE-Seq scoring matrix, you get a relative frequency of how often they cut, and these were included in that ONE-Seq matrix. We're seeing they cleave at almost similar efficiencies to the wild type sequence or actual target sequence. So it's another reason we're less concerned about these, but it's very important to be aware that they exist and to call that out as known as a possibility where you might have to come up with an alternate strategy or something.
Again, how close are all of your putative off-target sites to your on-target sites? We were fortunate we have two guide RNAs. None of our off-target effects are really possible off-targets. We're within five to 10 megabases of our actual targeted sites, giving us more confidence in the safety profiling of our guides.
Now if we go to the next slide, this is, really, the important part, when you actually look in the cell and you see what's going on. So this is our Amplicon sequencing for HSPC, our target cell type, treated with both gRNAs at the same time. We've gone through and looked at all of the sites, so it was all the tier 1, tier 2a and 2b. Because we had so few sites, we were actually able to additionally include hematopoietic-expressed tier 3 sites. These are normally excluded from analysis, but as you saw earlier, we had one that had only had 67 tier 1 sites identified total. So doing an amplicon sequencing of all that was not exactly difficult.
When we got down to it, all of those sites—we started out with almost 100,000 putative sites between two gRNAs—and we only had detection at eight sites. And all of those sites were at low percentage frequencies. The only one you can see here with the off-target 8 with a high concern score is actually not in a hematopoietic-expressed gene, so we have very high confidence that these will be well-tolerated as we go on and develop our therapeutic.
That was the small-scale RNA changes. Now if we look at the large scale and utilizing the SAFER Detection, there's a lot of really great data and ability in this. And so these were cells that, once again, were treated with both guide RNAs, so there is an expected rearrangement event where we excise that fraction of a gene and now create a new template. What you can see here in this blue bar is actually representative of things that might be what we were looking for, right? That excision. So you're getting a joining of the UP4 site to the DN4 site, and now all of the GAA expansion is gone.
That was detected readily in this assay, which was really a nice kind of sanity check that we're getting the results that would be expected in this. If we look for UP4- or DN4-specific rearrangement events, what we're seeing is nothing appears to be off-target mediated. They're very low numbers to begin with, and even those are more apparent to be maybe homology-mediated as opposed to off-target.
We tested the same thing orthogonally again, as you've heard many times—it's a common theme in safety testing. This is where we worked with KromaTiD utilizing some of their chromosome painting technologies. What you can see is when we look at events that are at our break sites, we aren't seeing anything large-scale different between what happens if we don't treat the cells and what happens when we do treat the cells. This really coincides with what we saw there in the SAFER Detection and really gives us a nice orthogonal view of how these could be very safe therapeutics. And as you know, that's part of the package we need to put together to get this IND in, is how safe is all of this? What you can see is if you look at the whole chromosome in an unbiased way by G-banding, we're also still seeing nothing of significant difference between their treated cells and the treated cells.
We've gone from very granular to very large scale, we're seeing an excellent safety profile, which is great for us, and we're seeing that all of this data is very complementary of each other, showing that the assays are functioning properly and giving us the kind of data we're going to need.
Our work with SeQure has been absolutely instrumental in defining off-target events and enhancing our safety site, so we can get this HSPC product out to patients that need it. And the whole of the data generated and demonstrating clean editing profiles is going to help advance this therapy towards IND application and into the clinical trials. It's really important to pick the right partners and look the right ways to identify your safety profiling so that we can make the best products available. We want to thank all of our collaborators.
Daniel Nguyen: Great. Thanks so much, Colin, Stephanie and Vijetha. We're moving into the Q&A session. My name's Daniel Lewin. I'm with MaxCyte and SeQure, and I’m going to help moderate this session.
The first question that we have looks like it's for the entire team. Does the off-target analysis results include only exons or does it also include introns?
Vijetha Vemulapalli: It does include introns as well. If you're thinking about the ONE-Seq assay, we look across the entire genome, so we're not going to exclude any sites, really. Even if it is in a repetitive region that's kind of hard to amplify, we can capture all of that in the ONE-Seq assay. Even with the other assays like GUIDE-Seq or DEUX-Seq, we would look across the genome regardless of exactly where the off-targets are present.
Colin Exline: I'll add that our results definitely included multiple intronic regions, multiple untranslated regions, multiple intergenic regions. So that’s all data that's captured.
Daniel Nguyen: Well, thank you, Colin, Vijetha, and thanks for the question. Colin, we do have another question here. What do you do with all of this off-target data?
Colin Exline: Data on its own is just data. You have to look into it and determine for yourself what the safety profiling is on a case by case basis almost. So as I said, our most concerning off-target that's been confirmed in cells is not in the hematopoietic expressed lineages. So clearly it's not causing rearrangements. Back to the previous question, it's actually an intron site, so you can kind of take that and go, alright, this is probably something we need to be aware of but is not causing a massive problem for our product.
One of the things we like to show is how everything lines up with how the numbers of putative off-target sites versus other known gRNAs, but on its own, that data doesn't mean much because you could have one putative off-target site but if it's in the exact wrong gene, you're not going to use that at all. So it's really a case by case basis. It builds a portfolio and kind of a whole view of it, and you just have to take that larger-scale and say, okay, what's important? Are these important, or do we just need to be aware of them and continue to monitor them in our further safety studies, in toxicology studies, and in patients. And really build that confidence that you're not affecting something that's going to be crucial for your downstream application.
Stephanie Cherqui: Yeah, I want to add that our clinical scale product, we will do testing, but we'll do with SeQure, to test really quickly all our product to make sure that this putative off-target will not be present or will not be an issue. So that will also be something we'll do.
Vijetha Vemulapalli: I think adding a little bit to what Colin and Stephanie said, at ASGCT there was a session where Kiran Musunuru was one of the panelists. He was involved in the treatment for baby Muldoon. It's been in the news, I'm sure all of you are aware for the rare liver disease. So the session was geared around discussing: how do you look at the off-target data to identify what the impact should be? A lot of the discussion came back to: what do you see in your cell type of interest, in your therapy? And also, what are all the functional annotations that you need to look at to identify what the impact of the off-target might be? All the annotation data that we provide is geared towards helping you make those decisions exactly. So you can look at the tiering, look at the characteristics of the off-targets, and based off of that, make informed decisions on what the risk of having that off-target might be.
Daniel Nguyen: Fantastic. Thank you all for answering that. We have a question specifically for Colin. With SAFER Detection, you detected homology-mediated translocations. Could you explain how you determine if a translocation is homology-mediated?
Colin Exline: So that actually might be more of a question for Vijetha because that's part of the report you get. What I would imagine is it's looking at the sequence and seeing: do you have these microhomology areas within x number of base pairs of the event, which would be indicative of a homology-mediated event? I don't think it's definitively something you can say, but it's most likely homology-mediated. What you can say is it was not off-target mediated because it wasn't between the on-target site and a predicted off-target site.
Vijetha Vemulapalli: Yeah, that's great Colin. It's a good explanation.
Daniel Nguyen: Excellent. A follow-up question: What strategies can be used to prevent Cas9 from rebinding to PAM sites?
Stephanie Cherqui: Maybe I will start answering this question. As I explain in our case is that we are using an RNP complex and that we electroporate—this is one of the safest strategies to use Cas9, as opposed to providing a vector with a long-term expression of Cas9. Because when you do that, the Cas9 stay about two days in the cell, so usually, they do their job and after they disappear. The chance that the Cas9 rebinding to, over time, sequence is much lower than when you express, long-term, the Cas9.
Daniel Nguyen: Absolutely. Stephanie, you’ve chosen Cas9 as the editor of choice, at least for this program. Are you thinking beyond Cas9, prime editors, whatnot? We've seen a rise in base editors as well from the SeQure front.
Stephanie Cherqui: Yeah, absolutely. I mean the technology keeps moving, keeps changing, so for other programs, for other diseases, we are using base editing. We just got our base editor designed by Alex Komor, which is one of the experts. But what I will do is before going through years of testing and work down the line, I will make sure that we work with SeQure to screen and make sure that our base editors are safe and with no worrisome off-target sites.
Daniel Nguyen: Thank you, Stephanie. Vijetha ,do you want to add any comments, at least in terms of what you've been seeing with the rise of base editors or other editing modalities?
Vijetha Vemulapalli: I think there's a lot of interest currently in moving to base-editing modalities. One of the things to consider is what assays are you going to use. Some of the nomination assays we have, especially the ONE-Seq assay, can be used for base editing as well. So just want to throw that out there because not too long ago we were Cas9 only, then we moved to Cas12 in addition to Cas9. But base editing and prime editing are the directions lots of people are moving in currently.
Daniel Nguyen: Thanks, Vijetha. We actually have a decent amount of international organizations who've joined the webinar. Vijetha, this is a question for you. Has SeQure worked with groups outside of the US and interacted with other regulatory bodies aside from the FDA?
Vijetha Vemulapalli: Yeah, we have worked with several clients that are in Europe. We can work with organizations anywhere in the world. I think it's worth highlighting the in silico products and the ONE-Seq. For some countries, it can be a challenge to send materials. With the ONE-Seq assay and the in silico products, we don't need the guide, we don't need any material at all, and we can still do the nomination assays for you. That's one.
To the second part of your question, we have engaged with some of the European regulatory bodies, with our clients filing with the European regulatory bodies, specifically around PEI. And we've done some enhancements to the products as well, to meet the requirements from these organizations. So we do have experience supporting IND filings with various regulatory bodies.
Daniel Nguyen: That's all the questions that we have today. Thank you everyone in the audience for joining us. Thank you, Stephanie, Colin, for participating in this webinar and presenting your work to the entire audience here today.