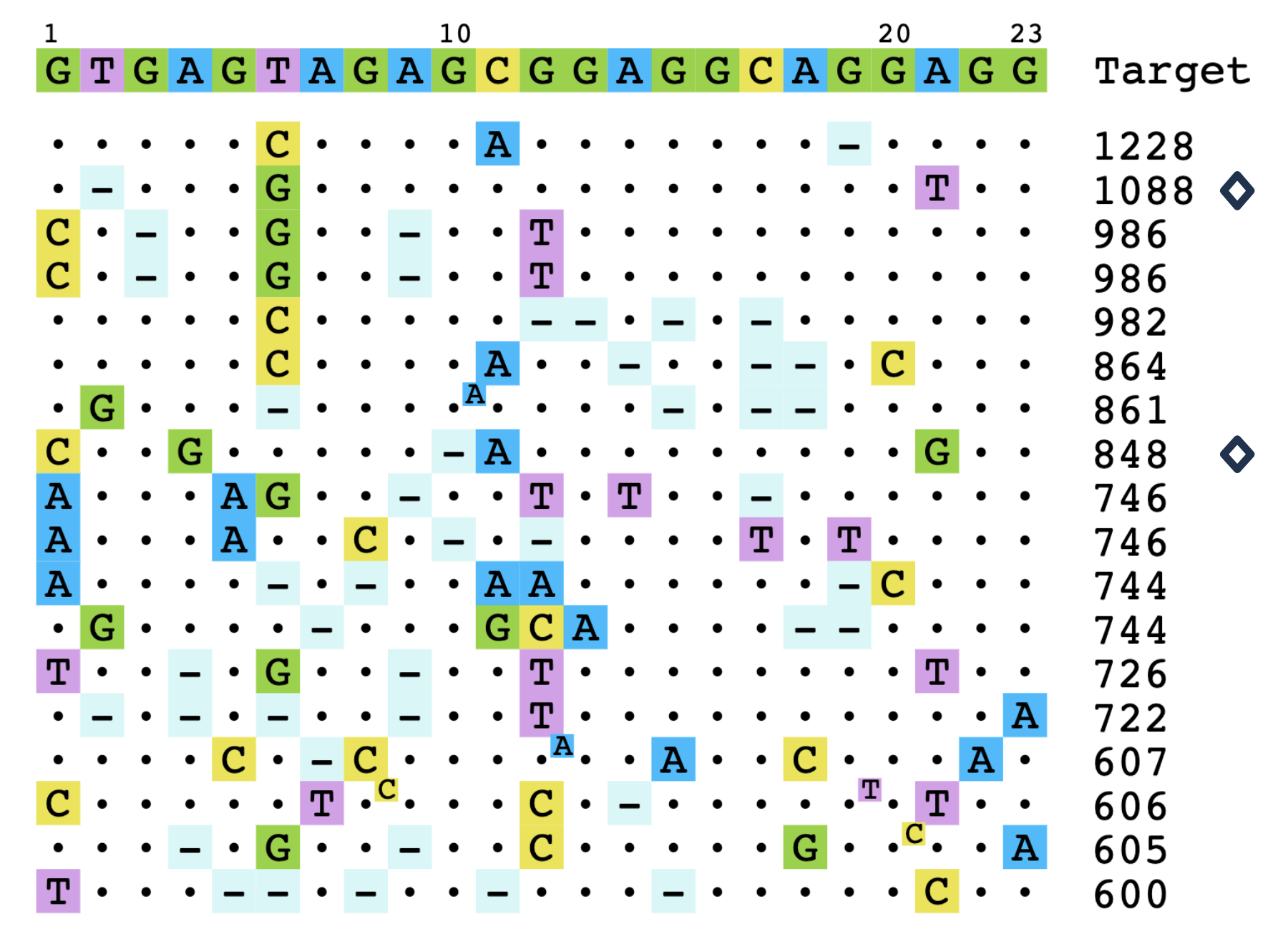
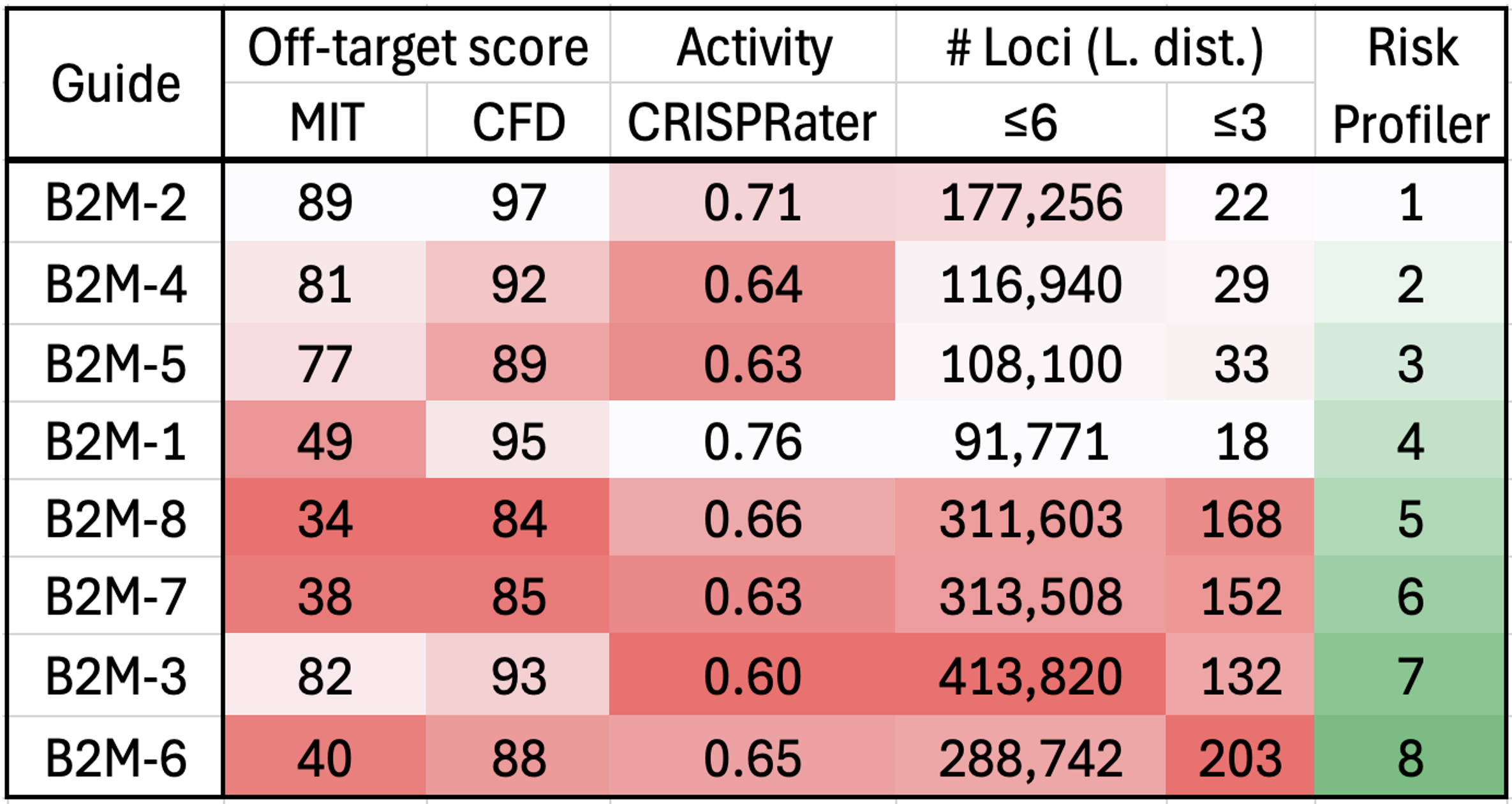
Comprehensive Suite of On-Target and Off-Target Assessment Assays
MaxCyte's SeQure is a comprehensive genotoxicity assessment platform that integrates screening, nomination and confirmation assays, providing unmatched precision in guide RNA selection and off-target risk assessment. By addressing safety and regulatory expectations early, our platform reduces development risks, streamlines therapeutic design, and accelerates your path to the clinic with confidence and efficiency.
A comprehensive portfolio of gene-editing assays
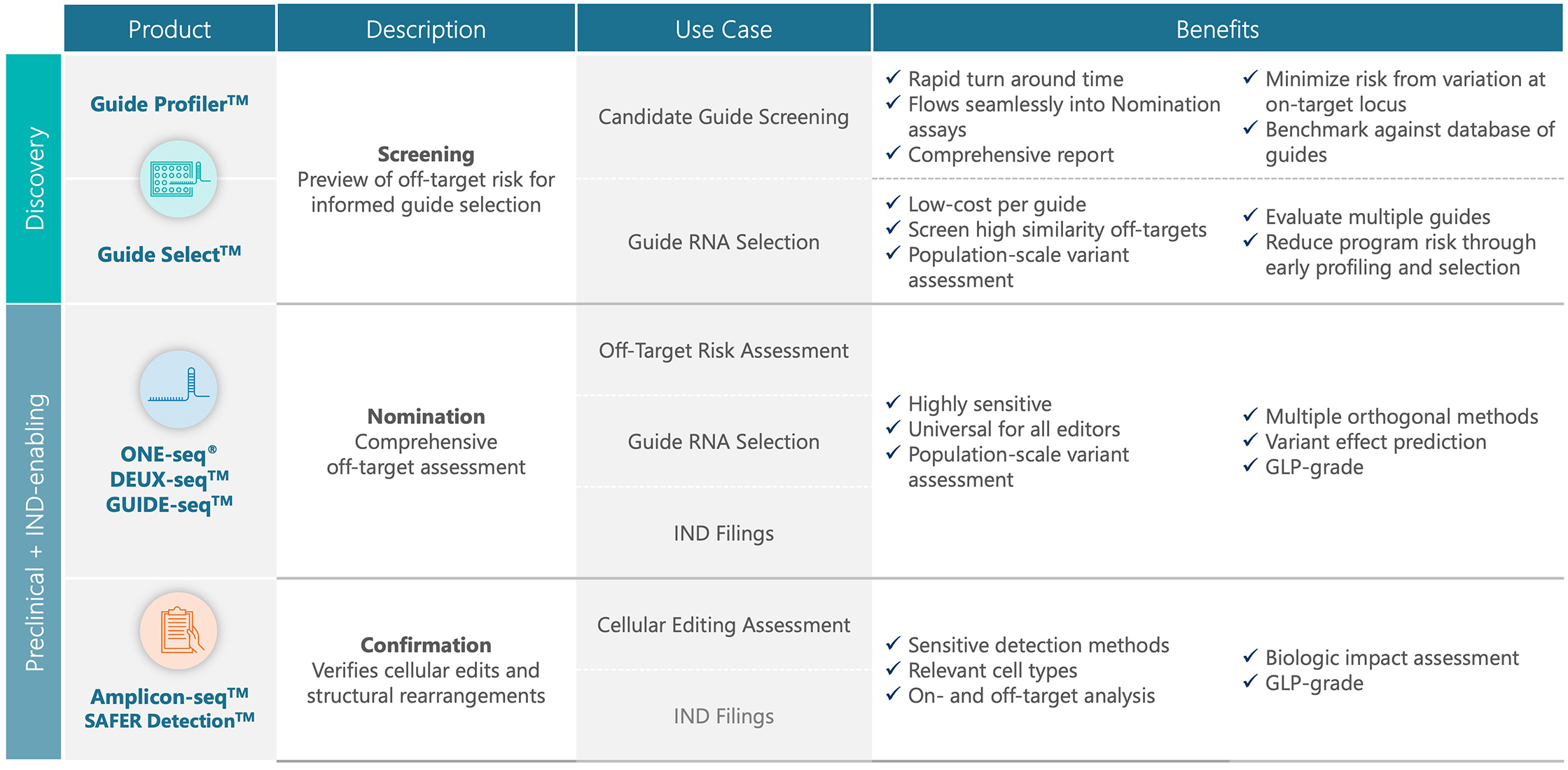
Screening assays
Preview of off-target risk for informed guide selection
Nomination assays
Comprehensive off-target assessment
Confirmation assays
Verifies cellular edits and structural rearrangements
Resources
Ready to learn more?