Scientific Brief
MaxCyte Electroporation Enables Efficient Antibody Discovery by Mammalian Display
Abstract
Therapeutic antibody developers constantly strive to find new ways to identify promising candidates and eliminate problematic ones earlier in the discovery and development process so that resources can be invested in only the best leads. Mammalian display identifies antibodies with desired antigen affinity and favorable biophysical properties, which may predict good developability1.
Identification of the best antibody candidates relies on screening large libraries in a workflow that ensures high sequence diversity and representation. In this pilot study, researchers transfected a subset of an IgG plasmid library to evaluate two electroporation methods in their mammalian display workflow. MaxCyte electroporation enabled better sequence diversity and library coverage with a streamlined workflow.
MaxCyte workflow for mammalian display
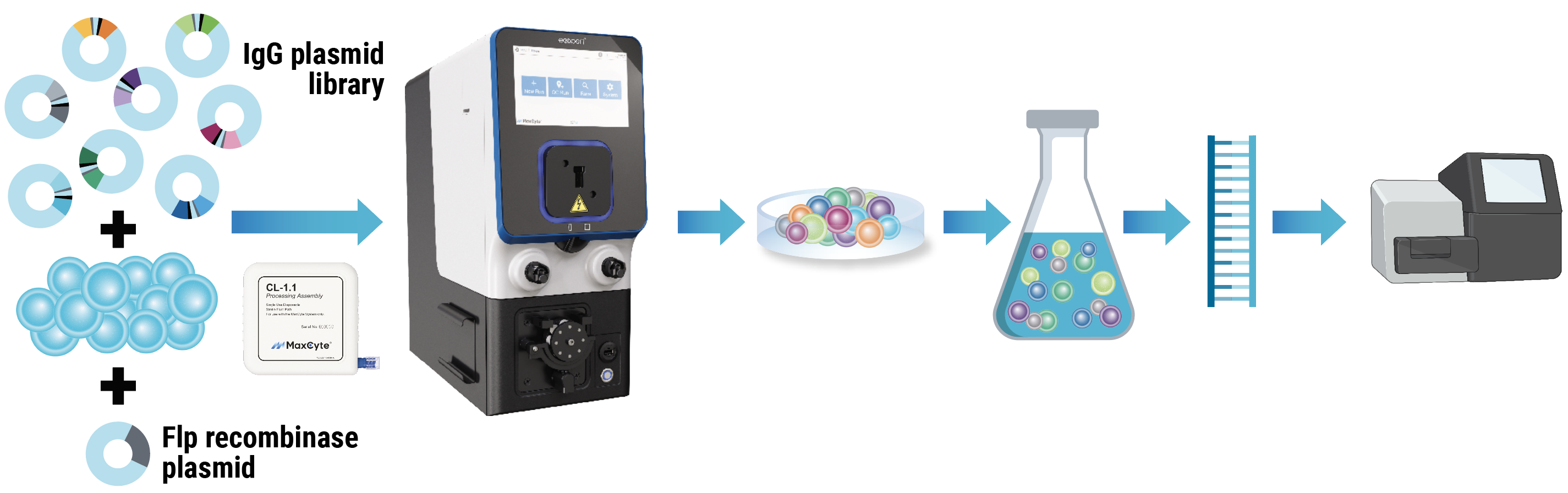
MaxCyte workflow. An engineered CHO cell line containing a landing pad for Flp recombinase-mediated targeted integration was cotransfected with a Flp recombinase plasmid and an IgG plasmid library encoding an antibiotic resistance gene. After resting, electroporated cells were cultured under antibiotic selection for nine days. Post-selection, 5 million cells were harvested, and mRNA was isolated, reverse-transcribed and sequenced.
Results
MaxCyte electroporation is scalable and reproducible, enabling better sequence diversity and library coverage with a more straightforward, faster process.
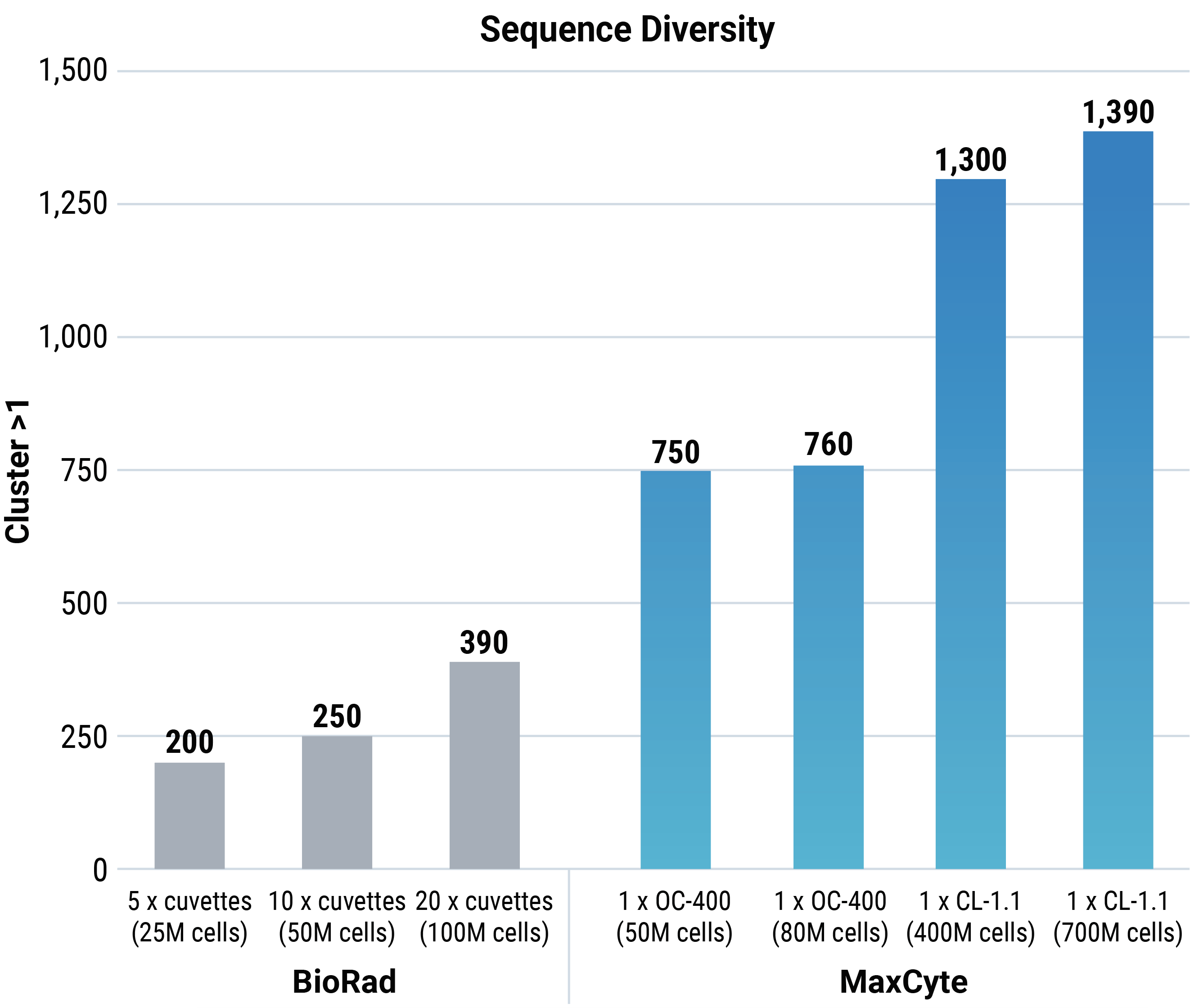
MaxCyte electroporation provided greater sequence diversity than Bio-Rad electroporation at every cell number tested. Furthermore, as the Bio-Rad method only transfects 5 million cells per cuvette, multiple rounds of transfection are needed per experiment. Conversely, MaxCyte electroporation enabled single processing assembly (cuvette) transfection at any cell number. Jaccard and Morisita analyses were done to evaluate the two transfection methods. As a control, the parental plasmid library was transformed into bacteria, and then DNA was recovered and sequenced.
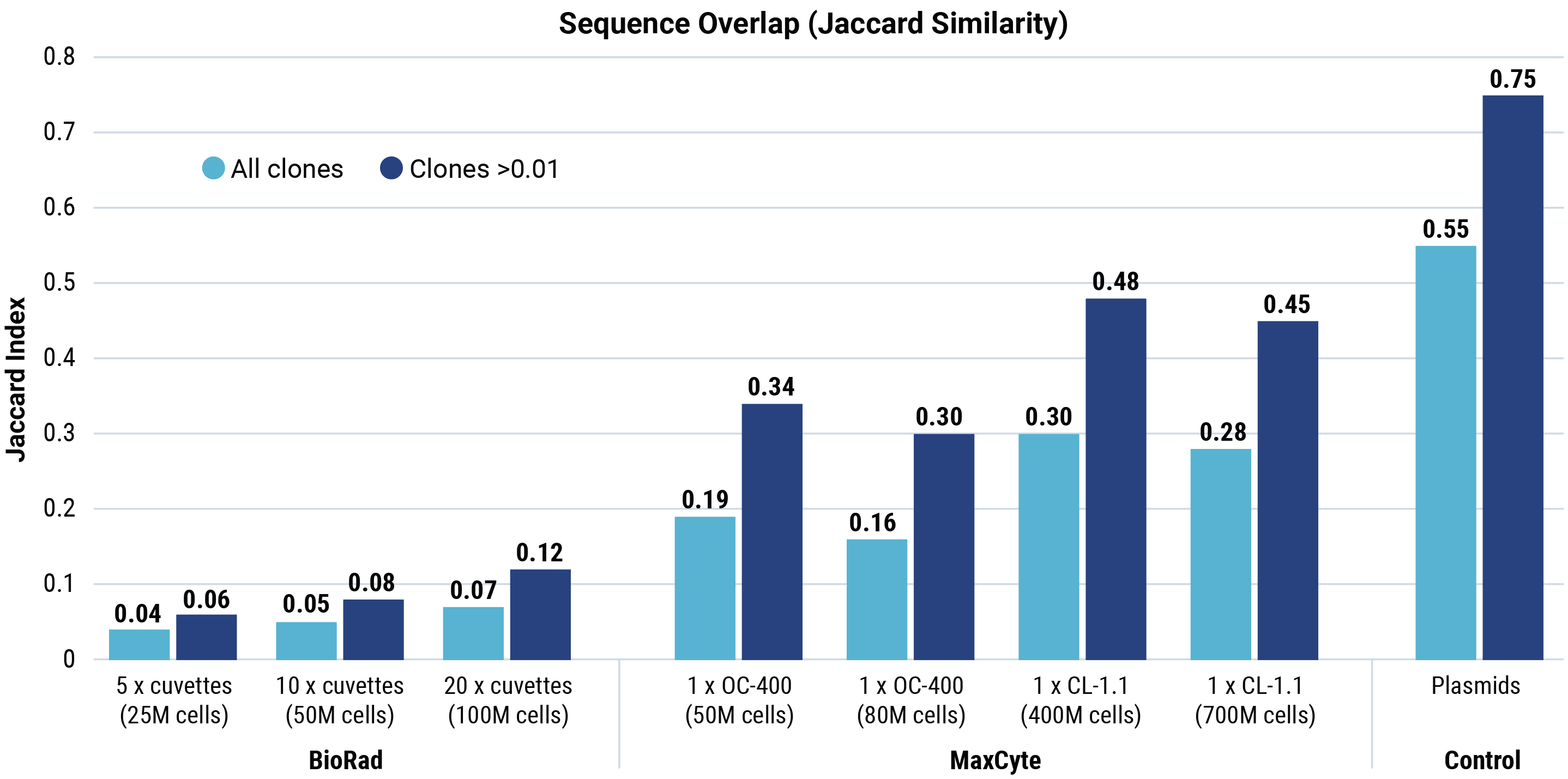
Jaccard similarity shows that for all conditions tested, cells generated by MaxCyte electroporation had more sequence overlap with the parental plasmid library than cells generated by Bio-Rad electroporation. A Morisita overlap score was calculated for each Bio-Rad and MaxCyte electroporation condition.
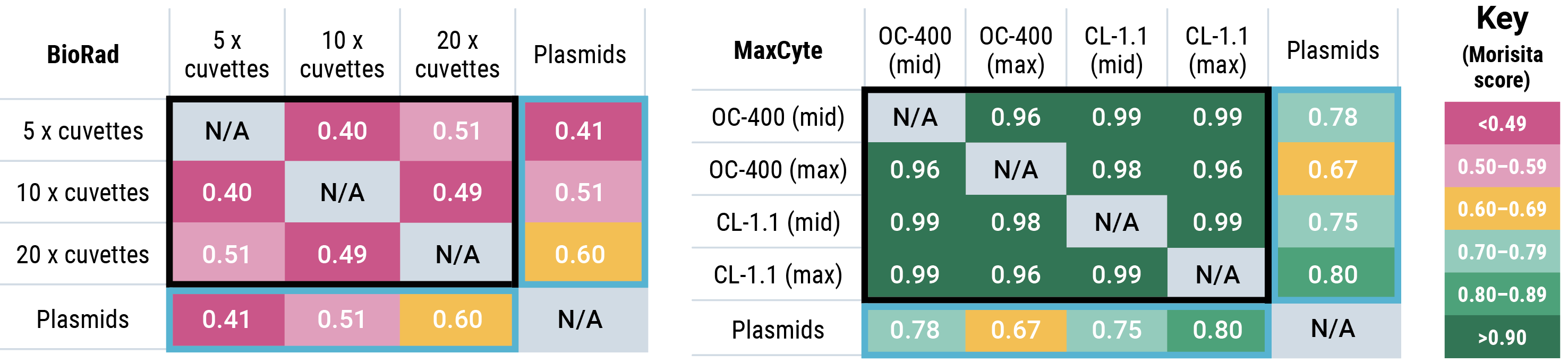
Morisita overlap scores with parental plasmids (light blue boxes) were higher for MaxCyte electroporation than for Bio-Rad, demonstrating that MaxCyte electroporation generated cells expressing sequences with more overlap with the parental plasmid library. The overlap between different transfections performed with each electroporation method (black boxes) was significantly lower with Bio-Rad electroporation (maximum 0.51) compared with MaxCyte electroporation (minimum 0.96), demonstrating the reproducibility of MaxCyte electroporation.
Summary
MaxCyte electroporation delivered greater sequence diversity and library coverage, enabling:
- A simplified workflow with one-shot electroporation of up to 700 million cells
- Reproducible transfection delivering consistent results
- Discovery of better antibody candidates using fewer resources and less hands-on time
MaxCyte’s Flow Electroporation technology can transfect up to 20 billion cells for mammalian display screening of large libraries for antibody discovery.
Related Resources
References
- Dyson, M. R., Masters, E., Pazeraitis, D., Perera, R. L., Syrjanen, J. L., Surade, S., … McCafferty, J. (2020). Beyond affinity: selection of antibody variants with optimal biophysical properties and reduced immunogenicity from mammalian display libraries. mAbs, 12(1). https://doi.org/10.1080/19420862.2020.1829335